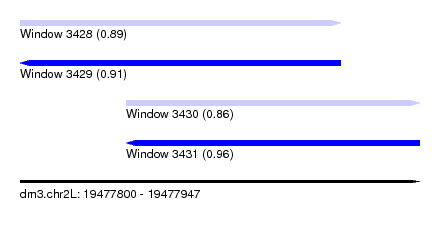
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,477,800 – 19,477,947 |
| Length | 147 |
| Max. P | 0.960299 |

| Location | 19,477,800 – 19,477,918 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.72 |
| Shannon entropy | 0.33876 |
| G+C content | 0.36656 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.46 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
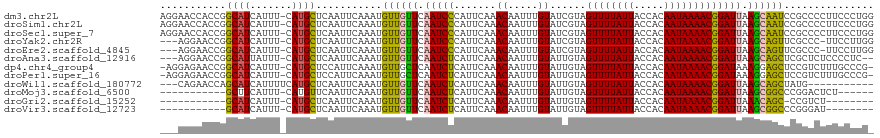
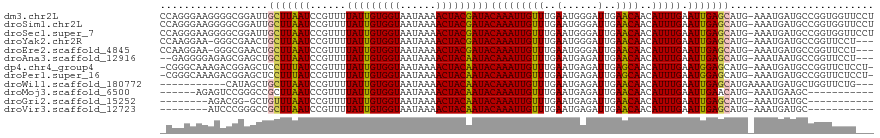
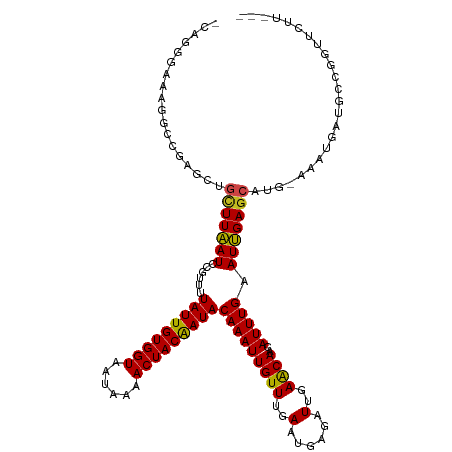
>dm3.chr2L 19477800 118 + 23011544 AGGAACCACCGGCAUCAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAAUCCGCCCCUUCCCUGG .((((....((((((.....-.))))...........((((((.(((((.......((.....))......((((((((.....))))))))))))).)))))).))....)))).... ( -20.00, z-score = -1.31, R) >droSim1.chr2L 19161459 118 + 22036055 AGGAACCACCGGCAUCAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAAUCCGCCCCUUCCCUGG .((((....((((((.....-.))))...........((((((.(((((.......((.....))......((((((((.....))))))))))))).)))))).))....)))).... ( -20.00, z-score = -1.31, R) >droSec1.super_7 3093003 118 + 3727775 AGGAACCACCGGCAUCAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAAUCCGCCCCUUCCCUGG .((((....((((((.....-.))))...........((((((.(((((.......((.....))......((((((((.....))))))))))))).)))))).))....)))).... ( -20.00, z-score = -1.31, R) >droYak2.chr2R 5975911 114 + 21139217 ---AGGAACCGGCAUCAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGUUCGCCC-UUCCUUGG ---(((((..(((..(((((-.(........).)))))(((((.(((((.......((.....))......((((((((.....))))))))))))).)))))...))).-)))))... ( -20.90, z-score = -1.68, R) >droEre2.scaffold_4845 17769846 114 - 22589142 ---AGGAACCGGCAUCAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGUUCGCCC-UUCCUUGG ---(((((..(((..(((((-.(........).)))))(((((.(((((.......((.....))......((((((((.....))))))))))))).)))))...))).-)))))... ( -20.90, z-score = -1.68, R) >droAna3.scaffold_12916 4079012 113 - 16180835 ---AGGAACCGGCAUUAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCUCGCUCUCCCCUC-- ---.(((...(((((.....-.)))))..........((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))......)))....-- ( -22.20, z-score = -2.69, R) >dp4.chr4_group4 585346 116 - 6586962 -AGGAGAACCGGCAUCAUUU-CAUGCUCCAUUCAAAUGUUGCUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUAAAGGAGCUCCGUCUUUGCCCG- -.((((..((((((.(((((-.(((...)))..))))).)))).......((((..(((((....))))).((((((((.....))))))))))))...))..))))...........- ( -24.40, z-score = -1.77, R) >droPer1.super_16 892648 116 + 1990440 -AGGAGAACCGGCAUCAUUU-CAUGCUCCAUUCAAAUGUUGCUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUAAAGGAGCUCCGUCUUUGCCCG- -.((((..((((((.(((((-.(((...)))..))))).)))).......((((..(((((....))))).((((((((.....))))))))))))...))..))))...........- ( -24.40, z-score = -1.77, R) >droWil1.scaffold_180772 7212119 105 + 8906247 ---CAGAACCAGCAUCAUUUUCAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCUAUG----------- ---..(((..(((((.......)))))...)))....((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))....----------- ( -20.80, z-score = -3.07, R) >droMoj3.scaffold_6500 29180533 101 + 32352404 -----------GCUUCAUUU-CAUGUUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCGGCCCGGACUCU------ -----------.........-..........((....((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))...))....------ ( -15.40, z-score = -0.92, R) >droGri2.scaffold_15252 4207551 98 + 17193109 -----------GCAUCAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAACAGC-CCGUCU-------- -----------((((.....-.))))...........((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))-......-------- ( -18.50, z-score = -3.75, R) >droVir3.scaffold_12723 663579 99 + 5802038 -----------GCAUCAUUU-CAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCGGCCCGGGAU-------- -----------((((.....-.))))...((((....((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))...))))-------- ( -19.00, z-score = -1.71, R) >consensus ___AAGAACCGGCAUCAUUU_CAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCUCGCCCUUUCCCUG_ ...........((((.......))))...........((((((.(((((.......((.....))......((((((((.....))))))))))))).))))))............... (-14.14 = -13.46 + -0.69)
| Location | 19,477,800 – 19,477,918 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.72 |
| Shannon entropy | 0.33876 |
| G+C content | 0.36656 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.07 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19477800 118 - 23011544 CCAGGGAAGGGGCGGAUUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUGAUGCCGGUGGUUCCU ...(((((.(((((((((....)))))))))(((((((((......)))))))))(((((((((..(......)..))))..))))).((((.((((.-.....))))))))..))))) ( -30.70, z-score = -1.61, R) >droSim1.chr2L 19161459 118 - 22036055 CCAGGGAAGGGGCGGAUUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUGAUGCCGGUGGUUCCU ...(((((.(((((((((....)))))))))(((((((((......)))))))))(((((((((..(......)..))))..))))).((((.((((.-.....))))))))..))))) ( -30.70, z-score = -1.61, R) >droSec1.super_7 3093003 118 - 3727775 CCAGGGAAGGGGCGGAUUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUGAUGCCGGUGGUUCCU ...(((((.(((((((((....)))))))))(((((((((......)))))))))(((((((((..(......)..))))..))))).((((.((((.-.....))))))))..))))) ( -30.70, z-score = -1.61, R) >droYak2.chr2R 5975911 114 - 21139217 CCAAGGAA-GGGCGAACUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUGAUGCCGGUUCCU--- ...(((((-.((((...((((((((......(((((((((......)))))))))(((((((((..(......)..))))..))))).))))))))..-......))))..)))))--- ( -28.00, z-score = -1.97, R) >droEre2.scaffold_4845 17769846 114 + 22589142 CCAAGGAA-GGGCGAACUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUGAUGCCGGUUCCU--- ...(((((-.((((...((((((((......(((((((((......)))))))))(((((((((..(......)..))))..))))).))))))))..-......))))..)))))--- ( -28.00, z-score = -1.97, R) >droAna3.scaffold_12916 4079012 113 + 16180835 --GAGGGGAGAGCGAGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUAAUGCCGGUUCCU--- --..(((..((((.....))))..)))....(((((((((......)))))))))....(((((..(......)..))))).....((((((.((((.-.....))))))))))..--- ( -29.40, z-score = -2.85, R) >dp4.chr4_group4 585346 116 + 6586962 -CGGGCAAAGACGGAGCUCCUUUAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAGCAACAUUUGAAUGGAGCAUG-AAAUGAUGCCGGUUCUCCU- -...........((((..((....((((((.(((((((((......)))))))))(((((((((..(......)..))))..)))))))))))((((.-.....)))).))..)))).- ( -31.50, z-score = -2.41, R) >droPer1.super_16 892648 116 - 1990440 -CGGGCAAAGACGGAGCUCCUUUAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAGCAACAUUUGAAUGGAGCAUG-AAAUGAUGCCGGUUCUCCU- -...........((((..((....((((((.(((((((((......)))))))))(((((((((..(......)..))))..)))))))))))((((.-.....)))).))..)))).- ( -31.50, z-score = -2.41, R) >droWil1.scaffold_180772 7212119 105 - 8906247 -----------CAUAGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUGAAAAUGAUGCUGGUUCUG--- -----------..((((.(((((((......(((((((((......)))))))))(((((((((..(......)..))))..))))).)))))))((....))...))))......--- ( -23.40, z-score = -2.25, R) >droMoj3.scaffold_6500 29180533 101 - 32352404 ------AGAGUCCGGGCCGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAACAUG-AAAUGAAGC----------- ------............((((..((.(((((((((((((......)))))))))(((((((((..(......)..))))..)))))....))))..)-)....))))----------- ( -17.20, z-score = -0.34, R) >droGri2.scaffold_15252 4207551 98 - 17193109 --------AGACGG-GCUGUUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUGAUGC----------- --------((((((-(........)))))))(((((((((......)))))))))(((((((((..(......)..))))..)))))......((((.-.....))))----------- ( -21.50, z-score = -2.20, R) >droVir3.scaffold_12723 663579 99 - 5802038 --------AUCCCGGGCCGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUG-AAAUGAUGC----------- --------..........(((((((......(((((((((......)))))))))(((((((((..(......)..))))..))))).)))))))...-.........----------- ( -21.10, z-score = -1.61, R) >consensus _CAGGGAAAGGCCGAGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUG_AAAUGAUGCCGGUUCUU___ ..................(((((((......(((((((((......)))))))))(((((((((..(......)..))))..))))).)))))))........................ (-19.54 = -19.07 + -0.48)
| Location | 19,477,839 – 19,477,947 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Shannon entropy | 0.51324 |
| G+C content | 0.37033 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -9.04 |
| Energy contribution | -8.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
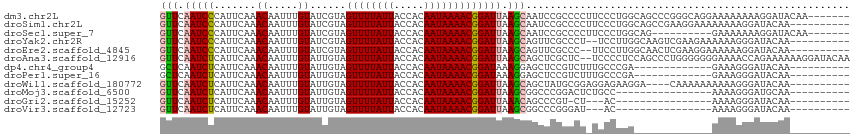
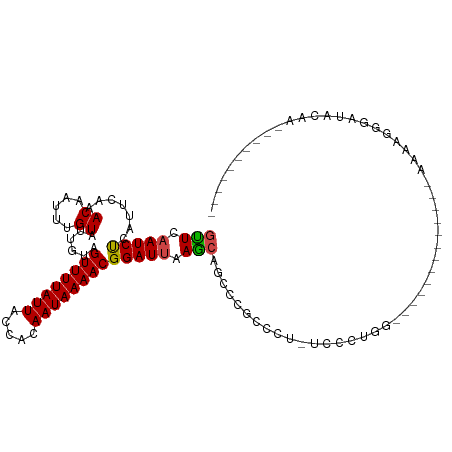
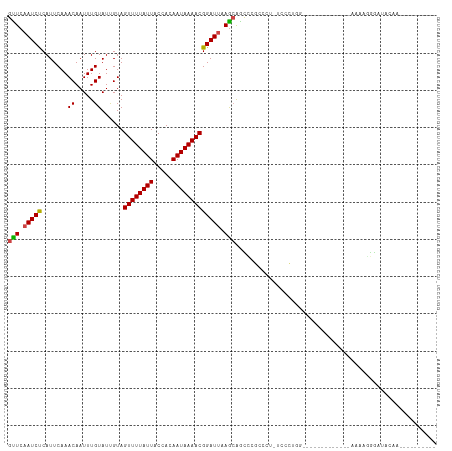
>dm3.chr2L 19477839 108 + 23011544 GUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAAUCCGCCCCUUCCCUGGCAGCCCGGGCAGGAAAAAAAAGGAUACAA------- .....................(((((((...((((((((.....))))))))(((((....)))))...(((.(((.((...)).))).))).........)))))))------- ( -25.10, z-score = -2.07, R) >droSim1.chr2L 19161498 105 + 22036055 GUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAAUCCGCCCCUUCCCUGGCAGCCGAAGGAAAAAAAAGGAUACAA---------- .....................(((((((...((((((((.....))))))))(((((....)))))...(((((..........))))).........)))))))---------- ( -20.10, z-score = -1.87, R) >droSec1.super_7 3093042 98 + 3727775 GUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAAUCCGCCCCUUCCCUGGCAG----------GAAAAAAAGGAUACAA------- .....................(((((((...((((((((.....))))))))(((((....)))))(((........)))..----------.........)))))))------- ( -19.50, z-score = -2.30, R) >droYak2.chr2R 5975947 103 + 21139217 GUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGUUCGCCCU--UCCUUGGCAAGUCGAAGAAAAAAGGGAUACAA---------- (((.(((((.......((.....))......((((((((.....))))))))))))).))).((...((((--((.((((....)))).))....))))..))..---------- ( -19.40, z-score = -1.57, R) >droEre2.scaffold_4845 17769882 103 - 22589142 GUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGUUCGCCC--UUCCUUGGCAACUCGAAGGAAAAAAGGAUACAA---------- .....................(((((((...((((((((.....))))))))(((((....)))))..((--(((...(....)..))))).......)))))))---------- ( -20.50, z-score = -2.06, R) >droAna3.scaffold_12916 4079048 113 - 16180835 GUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCUCGCUC--UCCCCUCCAGCCCUGGGGGGGAAAACCAGAAAAAAGGAUACAA .....................(((((((.(.((((((((.....))))))))((....(((.....))).--(((((((((....)))))))))...)).......).))))))) ( -30.60, z-score = -2.71, R) >dp4.chr4_group4 585384 92 - 6586962 GCUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUAAAGGAGCUCCGUCUUUGCCCGA-------------GAAAGGGAUACAA---------- .(((..((((((((..(((((....))))).((((((((.....))))))))))))...((.((.........))))))-------------))..)))......---------- ( -19.50, z-score = -1.78, R) >droPer1.super_16 892686 92 + 1990440 GCUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUAAAGGAGCUCCGUCUUUGCCCGA-------------GAAAGGGAUACAA---------- .(((..((((((((..(((((....))))).((((((((.....))))))))))))...((.((.........))))))-------------))..)))......---------- ( -19.50, z-score = -1.78, R) >droWil1.scaffold_180772 7212156 101 + 8906247 GUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCUAUGCGGAGGAGAAGGA----CAAAAAAAAAAGGGAUACAA---------- ((((..((((.(((....((((((((((((.((......)).)))))...)))))))..(((....)))))).)))).)))----)...................---------- ( -18.90, z-score = -2.25, R) >droMoj3.scaffold_6500 29180561 89 + 32352404 GUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCGGCCCGGACUCUGCC----------------AAAAGGGAUGCAA---------- ....(((((.......(((((....))))).((((((((.....)))))))))))))..(((.(((((......))----------------....))).)))..---------- ( -16.90, z-score = -0.68, R) >droGri2.scaffold_15252 4207579 85 + 17193109 GUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAACAGCCCGU-CU---AC----------------AAAAGGGAUACAA---------- (((.(((((.......(((((....))))).((((((((.....))))))))))))).)))..(((..-..---..----------------....)))......---------- ( -15.30, z-score = -2.43, R) >droVir3.scaffold_12723 663607 86 + 5802038 GUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCGGCCCGGGAU---AC----------------AAAAGGGAUACAA---------- (((.(((((.......(((((....))))).((((((((.....))))))))))))).)))(.(((.(...---.)----------------....)))...)..---------- ( -15.50, z-score = -1.20, R) >consensus GUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCCCGCCCU_UCCCUGG_____________AAAAGGGAUACAA__________ (((.(((((.......((.....))......((((((((.....))))))))))))).)))...................................................... ( -9.04 = -8.92 + -0.12)
| Location | 19,477,839 – 19,477,947 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.69 |
| Shannon entropy | 0.51324 |
| G+C content | 0.37033 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.27 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
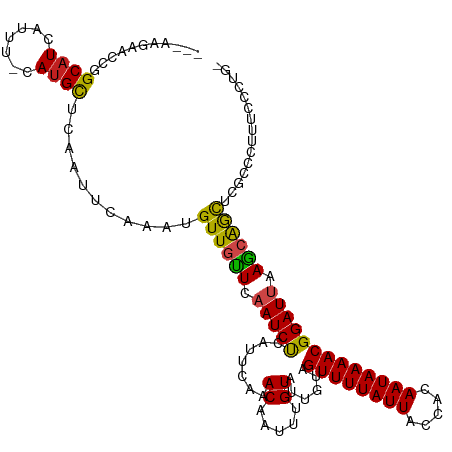
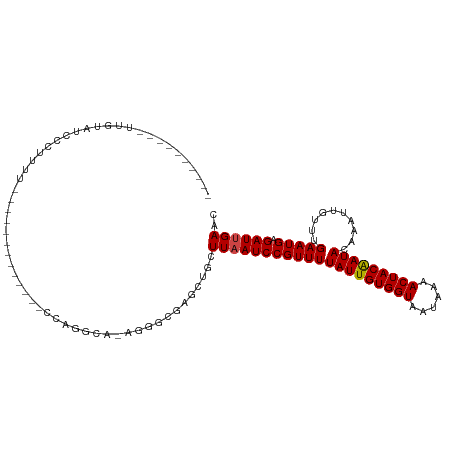
>dm3.chr2L 19477839 108 - 23011544 -------UUGUAUCCUUUUUUUUCCUGCCCGGGCUGCCAGGGAAGGGGCGGAUUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAAC -------....(((((.(((..((((.(((((....)).))).))))(((((((....)))))))..(((((((((......)))))))))..........))).)))))..... ( -29.70, z-score = -1.15, R) >droSim1.chr2L 19161498 105 - 22036055 ----------UUGUAUCCUUUUUUUUCCUUCGGCUGCCAGGGAAGGGGCGGAUUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAAC ----------..((((((.((((((((((..((...)))))))))))).))).)))(((((((....(((((((((......)))))))))(((.....)))....))))))).. ( -26.80, z-score = -1.39, R) >droSec1.super_7 3093042 98 - 3727775 -------UUGUAUCCUUUUUUUC----------CUGCCAGGGAAGGGGCGGAUUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAAC -------..((((((.(((((((----------((....))))))))).))).)))(((((((....(((((((((......)))))))))(((.....)))....))))))).. ( -27.30, z-score = -2.38, R) >droYak2.chr2R 5975947 103 - 21139217 ----------UUGUAUCCCUUUUUUCUUCGACUUGCCAAGGA--AGGGCGAACUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAAC ----------....(((((....(((..(((.(((....(((--.((((.....))))..)))....(((((((((......)))))))))))).)))...))).)))))..... ( -23.10, z-score = -1.40, R) >droEre2.scaffold_4845 17769882 103 + 22589142 ----------UUGUAUCCUUUUUUCCUUCGAGUUGCCAAGGAA--GGGCGAACUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAAC ----------....(((((...(((...(((.(((....(((.--((((.....))))..)))....(((((((((......)))))))))))).)))...))).)))))..... ( -21.40, z-score = -0.53, R) >droAna3.scaffold_12916 4079048 113 + 16180835 UUGUAUCCUUUUUUCUGGUUUUCCCCCCCAGGGCUGGAGGGGA--GAGCGAGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAAC ..............((.(((((((((.(((....))).)))))--)))).))....((((((((((((((((((((......)))))))))..........))))).)))))).. ( -35.30, z-score = -2.77, R) >dp4.chr4_group4 585384 92 + 6586962 ----------UUGUAUCCCUUUC-------------UCGGGCAAAGACGGAGCUCCUUUAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAGC ----------........(((..-------------(((((((((((((((.........)))))))(((((((((......)))))))))....))))))))..)))....... ( -24.30, z-score = -2.72, R) >droPer1.super_16 892686 92 - 1990440 ----------UUGUAUCCCUUUC-------------UCGGGCAAAGACGGAGCUCCUUUAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAGC ----------........(((..-------------(((((((((((((((.........)))))))(((((((((......)))))))))....))))))))..)))....... ( -24.30, z-score = -2.72, R) >droWil1.scaffold_180772 7212156 101 - 8906247 ----------UUGUAUCCCUUUUUUUUUUG----UCCUUCUCCUCCGCAUAGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAAC ----------....................----((..((((....(((....)))((((.(.(((((((((((((......))))))))).)))).).))))..))))..)).. ( -16.50, z-score = -1.93, R) >droMoj3.scaffold_6500 29180561 89 - 32352404 ----------UUGCAUCCCUUUU----------------GGCAGAGUCCGGGCCGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAAC ----------..((..(((....----------------(((...))).)))..))((((((((((((((((((((......)))))))))..........))))).)))))).. ( -18.40, z-score = -0.66, R) >droGri2.scaffold_15252 4207579 85 - 17193109 ----------UUGUAUCCCUUUU----------------GU---AG-ACGGGCUGUUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAAC ----------......(((.((.----------------..---.)-).)))..((((((((((((((((((((((......)))))))))..........))))).)))))))) ( -18.70, z-score = -2.06, R) >droVir3.scaffold_12723 663607 86 - 5802038 ----------UUGUAUCCCUUUU----------------GU---AUCCCGGGCCGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAAC ----------..((..(((....----------------..---.....)))..))((((((((((((((((((((......)))))))))..........))))).)))))).. ( -16.90, z-score = -1.38, R) >consensus __________UUGUAUCCCUUUU_____________CCAGGCA_AGGGCGAGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAAC ........................................................((((((((((((((((((((......)))))))))..........))))).)))))).. (-13.34 = -13.27 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:16 2011