| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,472,032 – 19,472,157 |
| Length | 125 |
| Max. P | 0.985598 |

| Location | 19,472,032 – 19,472,123 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.82 |
| Shannon entropy | 0.42623 |
| G+C content | 0.44323 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -15.41 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
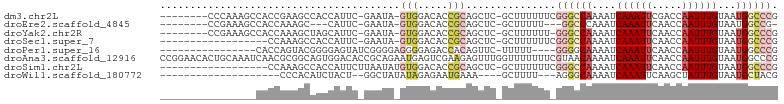
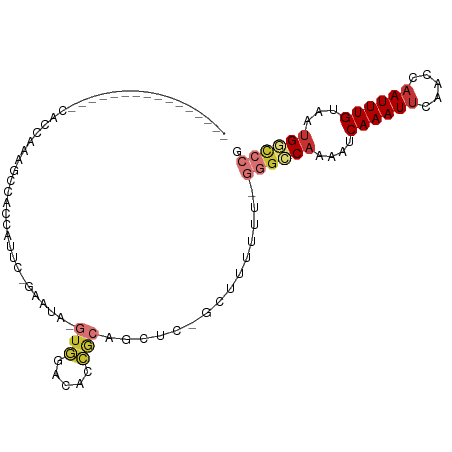
>dm3.chr2L 19472032 91 - 23011544 ---------------ACACCGCAGCUCGCUUUUUUCGGGCCAAAAUCAAAUUCGACCAAUUUGUAAUGGCCCGGAGUCG----------CAGGCG----AAGGCAAAUGUCUUCAUAUGC ---------------.....(((((..((...((((((((((....((((((.....))))))...))))))))))..)----------)..))(----(((((....))))))...))) ( -35.80, z-score = -4.33, R) >droEre2.scaffold_4845 17764239 87 + 22589142 ---------------ACACCGCAGCUCGCUUUUU---GGCCCAAAUCAAAUUCAACCAAUUUGUAAUGG-CCGGAGUCG----------CAGGCG----AAGGCAAAUGUCUUCAUAUGC ---------------.....(((((..((.((((---((((.....((((((.....))))))....))-))))))..)----------)..))(----(((((....))))))...))) ( -28.70, z-score = -2.74, R) >droYak2.chr2R 5970199 90 - 21139217 ---------------ACACCGCAGCUCGCUUUUUU-GGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCGGAGUCG----------CAGGCG----AAGGCAAAUGUCUUCAUUUGC ---------------.....(((((..((..((((-((((((....((((((.....))))))...))))))))))..)----------)..))(----(((((....))))))...))) ( -35.00, z-score = -4.33, R) >droSec1.super_7 3087394 91 - 3727775 ---------------ACACCGCAGCUCGCUUUUUUCGGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCGGAGUCG----------CAGGCG----AAGGCAAAUGUCUUCAUAUGC ---------------.....(((((..((...((((((((((....((((((.....))))))...))))))))))..)----------)..))(----(((((....))))))...))) ( -35.80, z-score = -4.83, R) >dp4.chr4_group4 578888 87 + 6586962 -------------------AGACCACAGUUCUUUUUGGGGCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCGGAAUUG----------AAGGAA----AAGGCAAAUGUCUUCAUAUGC -------------------...((.(((((((.....((((.....((((((.....)))))).....)))))))))))----------..))..----(((((....)))))....... ( -20.40, z-score = -0.82, R) >droPer1.super_16 886178 87 - 1990440 -------------------AGACCACAGUUCUUUUUGGGGCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCGGAAUUG----------AAGGAA----AAGGCAAAUGUCUUCAUAUGC -------------------...((.(((((((.....((((.....((((((.....)))))).....)))))))))))----------..))..----(((((....)))))....... ( -20.40, z-score = -0.82, R) >droAna3.scaffold_12916 4073164 105 + 16180835 ACACCGCAGAAUGAGUCGAAGAGUUUGGUUUUUUUCGUAACAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCGGAGU-G----------AAGGCA----GAGGCAAAUGUCUUCAUAUGU ...(((..............((((((((((((.........))))))))))))..(((........)))..))).((-(----------((((((----........))))))))).... ( -24.90, z-score = -1.09, R) >droSim1.chr2L 19153839 91 - 22036055 ---------------ACACCGCAGCUCGCUUUUUUCGGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCGGAGUCG----------CAGGCG----AAGGCAAAUGUCUUCAUAUGC ---------------.....(((((..((...((((((((((....((((((.....))))))...))))))))))..)----------)..))(----(((((....))))))...))) ( -35.80, z-score = -4.83, R) >droWil1.scaffold_180772 7201517 96 - 8906247 ------------------------AUGAAAGCUUUUAGGGCAAAAUCAAAUUCAAGCUAUUUGUAAUGCUACGGAAAUAGGAAUGGGCCAAGACAAGGCAAGGCAAAUGUCUUCAUAUGC ------------------------(((((.(((((...(((..(.((...(((.(((..........)))...)))....)).)..))).....)))))..(((....)))))))).... ( -17.20, z-score = 0.48, R) >consensus _______________ACACCGCAGCUCGCUUUUUUCGGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCGGAGUCG__________CAGGCG____AAGGCAAAUGUCUUCAUAUGC ................................((((((((((....((((((.....))))))...))))))))))................(((....(((((....)))))....))) (-15.41 = -16.30 + 0.89)


| Location | 19,472,065 – 19,472,157 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 65.66 |
| Shannon entropy | 0.65804 |
| G+C content | 0.46315 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -9.46 |
| Energy contribution | -10.55 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19472065 92 - 23011544 --------CCCAAAGCCACCGAAGCCACCAUUC-GAAUA-GUGGACACCGCAGCUC-GCUUUUUUCGGGCCAAAAUCAAAUUCGACCAAUUUGUAAUGGCCCG --------...(((((...((((.......)))-)....-((((...)))).....-)))))...(((((((....((((((.....))))))...))))))) ( -25.00, z-score = -2.51, R) >droEre2.scaffold_4845 17764272 85 + 22589142 --------CCGAAAGCCACCAAAGC---CAUUC-GAAUA-GUGGACACCGCAGCUC-GCUUUUU---GGCCCAAAUCAAAUUCAACCAAUUUGUAAUGGCCG- --------.(....)........((---(((((-((((.-((((...)))).....-(((....---)))..................))))).))))))..- ( -16.50, z-score = -0.45, R) >droYak2.chr2R 5970232 91 - 21139217 --------CCGAAAGCCACCAAAGCUAGCAUUC-GAAUA-GUGGACACCGCAGCUC-GCUUUUUU-GGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCG --------.(....).....(((((.(((((..-..)).-((((...)))).))).-)))))...-((((((....((((((.....))))))...)))))). ( -23.70, z-score = -1.96, R) >droSec1.super_7 3087427 82 - 3727775 ------------------CCAAAGCCACCAUUC-GAAUA-GUGGACACCGCAGCUC-GCUUUUUUCGGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCG ------------------...............-(((.(-(((..(......)..)-)))...)))((((((....((((((.....))))))...)))))). ( -21.30, z-score = -2.51, R) >droPer1.super_16 886211 82 - 1990440 ----------------CACCAGUACGGGGAGUAUCGGGGAGGGGAGACCACAGUUC-UUUUU----GGGGCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCG ----------------..((.((((.....)))).))....((....)).(((...-...))----)((((.....((((((.....)))))).....)))). ( -21.90, z-score = -0.55, R) >droAna3.scaffold_12916 4073196 103 + 16180835 CCGGAACACUGCAAAUCAACGCGGCAGUGGACACCGCAGAAUGAGUCGAAGAGUUUGGUUUUUUUCGUAACAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCG .(((..((.(((((((....((((.........)))).............((((((((((((.........)))))))))))).....))))))).))..))) ( -26.30, z-score = -1.23, R) >droSim1.chr2L 19153872 84 - 22036055 ------------------CCAAAGCCACCAUUCUUAAUAUGUGGACACCGCAGCUC-GCUUUUUUCGGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCG ------------------..(((((..............(((((...)))))....-)))))...(((((((....((((((.....))))))...))))))) ( -20.67, z-score = -2.45, R) >droWil1.scaffold_180772 7201564 74 - 8906247 --------------------CCCACAUCUACU--GGCUAUAUAGAGAAUGAAA----GCUUUU---AGGGCAAAAUCAAAUUCAAGCUAUUUGUAAUGCUACG --------------------.......(((..--((((..............)----)))..)---))((((....(((((.......)))))...))))... ( -9.84, z-score = 0.54, R) >consensus ________________CACCAAAGCCACCAUUC_GAAUA_GUGGACACCGCAGCUC_GCUUUUUU_GGGCCAAAAUCAAAUUCAACCAAUUUGUAAUGGCCCG ........................................(((.....)))...............((((((....((((((.....))))))...)))))). ( -9.46 = -10.55 + 1.09)
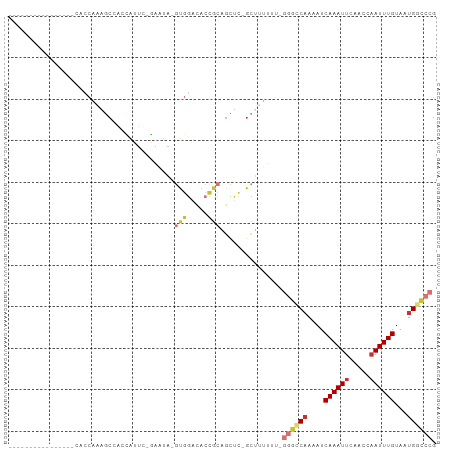
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:12 2011