
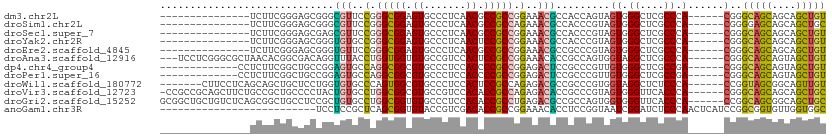
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,465,166 – 19,465,290 |
| Length | 124 |
| Max. P | 0.989599 |

| Location | 19,465,166 – 19,465,256 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.61025 |
| G+C content | 0.70720 |
| Mean single sequence MFE | -47.57 |
| Consensus MFE | -18.18 |
| Energy contribution | -17.64 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19465166 90 + 23011544 ---------------UCUUCGGGAGCGGGCGUUCCGGGCGGAGUGCCCUCAACGCCGCCGGAAACGCCACCAGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGU ---------------...(((((.((((((.((((((.(((.((.......)).)))))))))...((((.....)))))))))))).------)))(((((....))))) ( -42.90, z-score = -0.77, R) >droSim1.chr2L 19147744 90 + 22036055 ---------------UCUUCGGGAGCGGGCGUUCCGGGCGGAGUGCCCUCAACGCCGCCAGAAACGCCACCCGUAGUGGGCUCGCCCA------CGGGGAGCAGCAGCUGC ---------------((((((((....((((((.(.(((((.((.......)).))))).).)))))).)))...(((((....))))------))))))((((...)))) ( -43.20, z-score = -0.73, R) >droSec1.super_7 3080522 90 + 3727775 ---------------UCUUCGGGAGCGAGCGUUCCGGGCGGAGUGCCCUCAACGCCGCCGGAAACGCCACCCGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGU ---------------...(((((.((((((.((((((.(((.((.......)).)))))))))...((((.....)))))))))))).------)))(((((....))))) ( -43.80, z-score = -1.23, R) >droYak2.chr2R 5963296 90 + 21139217 ---------------UCUUCGGGAGCGGGUGUGCCGGGCGGAGUGCCCUCAACUCCGCCGGAAACGCCACCCGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGU ---------------((....)).(((((((.((..((((((((.......))))))))(....)))))))))).(((((....))))------)..(((((....))))) ( -50.30, z-score = -2.59, R) >droEre2.scaffold_4845 17757341 90 - 22589142 ---------------UCUUCGGGAGCGGGUGUUCCGGGCGGAGUGCCCUCAACGCCGCCGGAAACGCCGCCCGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGU ---------------...(((((.(((((((((((((.(((.((.......)).))))))).))))))((((.....)))).))))).------)))(((((....))))) ( -44.90, z-score = -0.73, R) >droAna3.scaffold_12916 4066736 102 - 16180835 ---UCCUCGGGCGCUAACACGGCGACAGGUUUACCUGGUGGUGUGCCGUCCACUCCGCCGGAAACACCGCCAGUGGUAGGCUCGCCCA------CGGGCAGCAGUAGCUGU ---.((..(((((((.....)))....(((((((((((((((((....(((........))).)))))))))..)))))))).)))).------.))(((((....))))) ( -45.80, z-score = -1.17, R) >dp4.chr4_group4 571880 92 - 6586962 -------------CCUCUUCGGCUGCCGGAGUGCCAGGCGGCGUGCCCUCCACCCCGCCGGAGACUCCGCCCGUUGUGGGCUCGCCGA------CGGGCAGCAGUAGCUGU -------------......(((((((.(((((.((.(((((.(((.....))).)))))))..)))))((((((((((....)).)))------)))))....))))))). ( -48.20, z-score = -1.71, R) >droPer1.super_16 879166 92 + 1990440 -------------CCUCUUCGGCUGCCGGAGUGCCAGGCGGCGUGCCCUCCACCCCGCCGGAGACUCCGCCCGUUGUGGGCUCGCCGA------CGGGCAGCAGUAGCUGU -------------......(((((((.(((((.((.(((((.(((.....))).)))))))..)))))((((((((((....)).)))------)))))....))))))). ( -48.20, z-score = -1.71, R) >droWil1.scaffold_180772 7190444 98 + 8906247 -------CUUCCUCAGCAGCUGCUCCUGGUGUGCCCAGUGGCGUGCCCUCCACUCCGCCAGAGACGCCGCCCGUGGUAGGCUCUCCCA------CCGGUAGCGGCAGUUGU -------......((((.((((((.((((((......((((.(((.....))).))))..((((.((((((...))).))))))).))------)))).)))))).)))). ( -41.70, z-score = -1.48, R) >droVir3.scaffold_12723 647268 104 + 5802038 -CCGCCGCAGCUUCUGCCGCUGCCCCUACUGUGCCUGGCGGCGUGCCGUCCACACCGCCAGAGACACCGCCCGUAGUGGGUUCACCCA------CGGGCAGCAGCAGCUGC -.....((((((...((.(((((((.....(((((((((((.(((.....))).))))))).).)))........(((((....))))------)))))))).)))))))) ( -56.50, z-score = -3.81, R) >droGri2.scaffold_15252 4193055 105 + 17193109 GCGGCUGCUGUCUCAGCGGCUGCCUCCGCUGUGCCUGGCGGUGUGCCCUCCACACCGCCUGAGACGCCGCCAGUGGUGGGUUCACCCA------CCGGCAGCGGCAGCUGC (((((((((((((((((((......)))))......(((((((((.....))))))))).)))))((.(((...((((((....))))------))))).))))))))))) ( -66.90, z-score = -4.33, R) >anoGam1.chr3R 4533607 85 - 53272125 --------------------------UCCUCCGCUCAGCGGUGUACCGUCGACACCGCCGGAAACACCUCCGGUAAUCGGAUCUCCCAACUCAUCCGGCGGUGUUGGUGGC --------------------------..(.((((...)))).)..((..(((((((((((((...(((...)))....((.....))......)))))))))))))..)). ( -38.40, z-score = -3.45, R) >consensus _______________UCUUCGGCAGCGGGUGUGCCGGGCGGAGUGCCCUCCACGCCGCCGGAAACGCCGCCCGUAGUGGGCUCGCCCA______CGGGCAGCAGCAGCUGU .............................(((..(.(((((.((.......)).))))).)..)))..(((.......)))................(((((....))))) (-18.18 = -17.64 + -0.54)


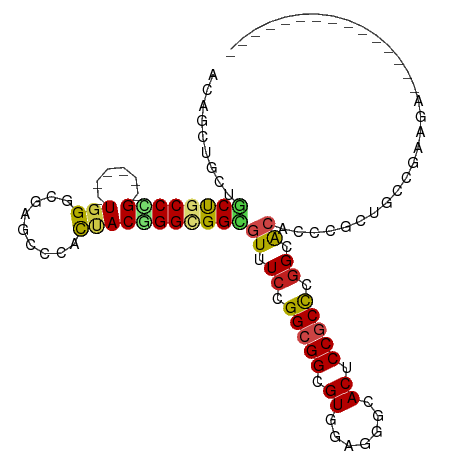
| Location | 19,465,166 – 19,465,256 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.61025 |
| G+C content | 0.70720 |
| Mean single sequence MFE | -48.18 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.75 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19465166 90 - 23011544 ACAGCUGCUGCUGCCCG------UGGGCGAGCCCACUACUGGUGGCGUUUCCGGCGGCGUUGAGGGCACUCCGCCCGGAACGCCCGCUCCCGAAGA--------------- ...((((.(((..((.(------((((....)))))....))..)))....))))((((((..((((.....))))..))))))............--------------- ( -40.50, z-score = -0.34, R) >droSim1.chr2L 19147744 90 - 22036055 GCAGCUGCUGCUCCCCG------UGGGCGAGCCCACUACGGGUGGCGUUUCUGGCGGCGUUGAGGGCACUCCGCCCGGAACGCCCGCUCCCGAAGA--------------- ((((...)))).....(------((((....)))))..((((.((((((((.(((((.((.......)).))))).))))))))....))))....--------------- ( -43.70, z-score = -0.85, R) >droSec1.super_7 3080522 90 - 3727775 ACAGCUGCUGCUGCCCG------UGGGCGAGCCCACUACGGGUGGCGUUUCCGGCGGCGUUGAGGGCACUCCGCCCGGAACGCUCGCUCCCGAAGA--------------- .((((....))))...(------((((....)))))..((((.((((((((.(((((.((.......)).))))).))))))))....))))....--------------- ( -43.60, z-score = -0.86, R) >droYak2.chr2R 5963296 90 - 21139217 ACAGCUGCUGCUGCCCG------UGGGCGAGCCCACUACGGGUGGCGUUUCCGGCGGAGUUGAGGGCACUCCGCCCGGCACACCCGCUCCCGAAGA--------------- .((((....))))...(------((((....)))))..((((.((((...((((((((((.......)))))).))))......))))))))....--------------- ( -41.60, z-score = -0.54, R) >droEre2.scaffold_4845 17757341 90 + 22589142 ACAGCUGCUGCUGCCCG------UGGGCGAGCCCACUACGGGCGGCGUUUCCGGCGGCGUUGAGGGCACUCCGCCCGGAACACCCGCUCCCGAAGA--------------- ..(((....((((((((------((((....)))))...)))))))(((((.(((((.((.......)).))))).)))))....)))........--------------- ( -41.70, z-score = -0.40, R) >droAna3.scaffold_12916 4066736 102 + 16180835 ACAGCUACUGCUGCCCG------UGGGCGAGCCUACCACUGGCGGUGUUUCCGGCGGAGUGGACGGCACACCACCAGGUAAACCUGUCGCCGUGUUAGCGCCCGAGGA--- .((((....)))).((.------((((((...(((.(((.((((((((..(((..........))).)))))..((((....))))..)))))).))))))))).)).--- ( -43.50, z-score = -0.40, R) >dp4.chr4_group4 571880 92 + 6586962 ACAGCUACUGCUGCCCG------UCGGCGAGCCCACAACGGGCGGAGUCUCCGGCGGGGUGGAGGGCACGCCGCCUGGCACUCCGGCAGCCGAAGAGG------------- .((((....)))).((.------(((((..((((.....))))(((((.((.(((((.(((.....))).))))).)).)))))....)))))...))------------- ( -49.50, z-score = -1.65, R) >droPer1.super_16 879166 92 - 1990440 ACAGCUACUGCUGCCCG------UCGGCGAGCCCACAACGGGCGGAGUCUCCGGCGGGGUGGAGGGCACGCCGCCUGGCACUCCGGCAGCCGAAGAGG------------- .((((....)))).((.------(((((..((((.....))))(((((.((.(((((.(((.....))).))))).)).)))))....)))))...))------------- ( -49.50, z-score = -1.65, R) >droWil1.scaffold_180772 7190444 98 - 8906247 ACAACUGCCGCUACCGG------UGGGAGAGCCUACCACGGGCGGCGUCUCUGGCGGAGUGGAGGGCACGCCACUGGGCACACCAGGAGCAGCUGCUGAGGAAG------- ......((((((...((------((((....))))))...)))))).(((..(((((.(((.....)))((..((((.....))))..))..)))))..)))..------- ( -44.60, z-score = -1.14, R) >droVir3.scaffold_12723 647268 104 - 5802038 GCAGCUGCUGCUGCCCG------UGGGUGAACCCACUACGGGCGGUGUCUCUGGCGGUGUGGACGGCACGCCGCCAGGCACAGUAGGGGCAGCGGCAGAAGCUGCGGCGG- (((((((((((((((((------((((....))))).....((.((((..(((((((((((.....))))))))))))))).))..))))))))))...)))))).....- ( -69.70, z-score = -5.04, R) >droGri2.scaffold_15252 4193055 105 - 17193109 GCAGCUGCCGCUGCCGG------UGGGUGAACCCACCACUGGCGGCGUCUCAGGCGGUGUGGAGGGCACACCGCCAGGCACAGCGGAGGCAGCCGCUGAGACAGCAGCCGC ((.(((((.((((((((------((((....))))))...))))))(((((.(((((((((.....)))))))))......(((((......)))))))))).))))).)) ( -69.30, z-score = -4.77, R) >anoGam1.chr3R 4533607 85 + 53272125 GCCACCAACACCGCCGGAUGAGUUGGGAGAUCCGAUUACCGGAGGUGUUUCCGGCGGUGUCGACGGUACACCGCUGAGCGGAGGA-------------------------- .((.((((((((.((((...((((((.....)))))).)))).))))))..((((((((((....).)))))))))...)).)).-------------------------- ( -41.00, z-score = -3.19, R) >consensus ACAGCUGCUGCUGCCCG______UGGGCGAGCCCACUACGGGCGGCGUUUCCGGCGGCGUGGAGGGCACUCCGCCCGGCACACCCGCUGCCGAAGA_______________ .........((((((((......((((....))))...))))))))((.((.(((((.((.......)).))))).)).)).............................. (-26.34 = -26.75 + 0.41)

| Location | 19,465,182 – 19,465,290 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.53487 |
| G+C content | 0.66572 |
| Mean single sequence MFE | -45.17 |
| Consensus MFE | -19.77 |
| Energy contribution | -19.46 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19465182 108 + 23011544 UUCCGGGCGGAGUGCCCUCAACGCCGCCGGAAACGCCACCAGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGUCCGGCUCAGACUUACGCACACCACUAUGCGAGGA------ ((((((.(((.((.......)).)))))))))..(((.......(((((....))))------)(((((((....)))))))))).....(((.((((........))))))).------ ( -49.00, z-score = -2.16, R) >droSim1.chr2L 19147760 108 + 22036055 UUCCGGGCGGAGUGCCCUCAACGCCGCCAGAAACGCCACCCGUAGUGGGCUCGCCCA------CGGGGAGCAGCAGCUGCCCGGCUCAGACUUACGCACACCACUAUGCCAGGA------ .(((.(((((.((.......)).)))))......(((.......(((((....))))------)(((.(((....))).))))))..........(((........)))..)))------ ( -41.50, z-score = -0.11, R) >droSec1.super_7 3080538 108 + 3727775 UUCCGGGCGGAGUGCCCUCAACGCCGCCGGAAACGCCACCCGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGUCCGGCUCAGACUUACGCACACCACUAUGCCAGGA------ ((((((.(((.((.......)).)))))))))..(((.......(((((....))))------)(((((((....)))))))))).....(((..(((........))).))).------ ( -45.00, z-score = -0.97, R) >droYak2.chr2R 5963312 108 + 21139217 UGCCGGGCGGAGUGCCCUCAACUCCGCCGGAAACGCCACCCGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGUCCGGCUCAGACUUACGCACACCACUAUGCCAGGA------ ..((.((((((((.......))))))))(....)(((.......(((((....))))------)(((((((....))))))))))..........(((........)))..)).------ ( -48.30, z-score = -1.71, R) >droEre2.scaffold_4845 17757357 108 - 22589142 UUCCGGGCGGAGUGCCCUCAACGCCGCCGGAAACGCCGCCCGUAGUGGGCUCGCCCA------CGGGCAGCAGCAGCUGUCCGGCUCAGACUUACGCACACCACUAUGCCAGGA------ .(((((((((...((((.(.(((.((.((....)).))..))).).)))))))))).------.(((((((....)))))))(((..((..............))..))).)))------ ( -46.64, z-score = -0.96, R) >droAna3.scaffold_12916 4066764 108 - 16180835 UACCUGGUGGUGUGCCGUCCACUCCGCCGGAAACACCGCCAGUGGUAGGCUCGCCCA------CGGGCAGCAGUAGCUGUCCGGCCCAGACAUAUGCCCACCAUUAUUCCCGGA------ ..((.((((((((....(((........))).))))))))((((((.(((((.....------((((((((....)))))))).....)).....))).))))))......)).------ ( -46.10, z-score = -2.17, R) >droWil1.scaffold_180772 7190468 114 + 8906247 UGCCCAGUGGCGUGCCCUCCACUCCGCCAGAGACGCCGCCCGUGGUAGGCUCUCCCA------CCGGUAGCGGCAGUUGUCCAGCCCAGACCUAUGCCCAUCAUUAUGCACAUGCCCGGA ......((((.(((.....))).))))..((((.((((((...))).)))))))...------((((..(.((((...(((.......)))...)))))........((....)))))). ( -33.80, z-score = 1.04, R) >droVir3.scaffold_12723 647298 103 + 5802038 UGCCUGGCGGCGUGCCGUCCACACCGCCAGAGACACCGCCCGUAGUGGGUUCACCCA------CGGGCAGCAGCAGCUGCCCCAGCCAGGCAUAUGCGCAUCAUUACCA----------- ((((((((((.(((.....))).))))))).......(((....(((((....))))------)(((((((....)))))))......)))......))).........----------- ( -48.70, z-score = -3.33, R) >droMoj3.scaffold_6500 29162376 103 + 32352404 UACCUGUUGGCGUGCCCUCCACUCCACCGGAGACGCCGCCCGUGGUGGGCUCCCCAA------ACGGCAGCAGCAGCUGCCCCAGCCAGGCAUAUGCCCAUCAUUAUCA----------- ........((((.((.((((........))))..)))))).(((((((((.......------..((((((....))))))((.....)).....))))))))).....----------- ( -40.50, z-score = -1.78, R) >droGri2.scaffold_15252 4193086 103 + 17193109 UGCCUGGCGGUGUGCCCUCCACACCGCCUGAGACGCCGCCAGUGGUGGGUUCACCCA------CCGGCAGCGGCAGCUGCCCCAGUCAGGCCUAUGCCCAUCAUUACCA----------- .(((((((((((((.....)))))))))...(((.........((((((....))))------))((((((....))))))...)))))))..................----------- ( -50.80, z-score = -3.13, R) >anoGam1.chr3R 4533617 99 - 53272125 -----AGCGGUGUACCGUCGACACCGCCGGAAACACCUCCGGUAAUCGGAUCUCCCAACUCAUCCGGCGGUGUUGGUGGCACCGGU-GGAGUCGUGAGCGGCGGU--------------- -----..(((((..((..(((((((((((((...(((...)))....((.....))......)))))))))))))..)))))))..-...((((....))))...--------------- ( -46.50, z-score = -1.75, R) >consensus UGCCGGGCGGAGUGCCCUCAACGCCGCCGGAAACGCCGCCCGUAGUGGGCUCGCCCA______CGGGCAGCAGCAGCUGUCCGGCUCAGACUUAUGCACACCACUAUGCCAGGA______ .....(((((.((.......)).)))))......(((((.....)).)))...............((((((....))))))....................................... (-19.77 = -19.46 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:10 2011