| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,820,760 – 1,820,854 |
| Length | 94 |
| Max. P | 0.681738 |

| Location | 1,820,760 – 1,820,854 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.57 |
| Shannon entropy | 0.27393 |
| G+C content | 0.44266 |
| Mean single sequence MFE | -18.81 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.35 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
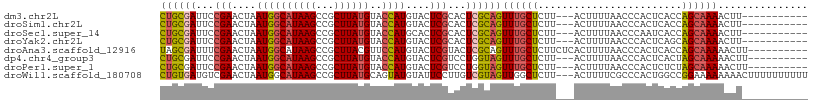
>dm3.chr2L 1820760 94 - 23011544 CUGCGAUUCCGAACUAAUGGCAUAAGCCGCUUAUGUACCAUGUACUCGCACUCGCAGUUUGCUCUU---ACUUUUAACCCACUCACCAGCAAAACUU----------- ((((((...(((....((((((((((...))))))..))))....)))...))))))((((((...---..................))))))....----------- ( -20.70, z-score = -2.69, R) >droSim1.chr2L 1804936 94 - 22036055 CUGCGAUUCCGAACUAAUGGCAUAAGCCGCUUAUGUACCAUGUACUCGCACUCGCAGUUUGCUCUU---ACUUUUAACCCACUCACCAGCAAAACUU----------- ((((((...(((....((((((((((...))))))..))))....)))...))))))((((((...---..................))))))....----------- ( -20.70, z-score = -2.69, R) >droSec1.super_14 1767466 94 - 2068291 CUGCGAUUCCGAACUAAUGGCAUAAGCCGCUUAUGUACCAUGCACUCGCACUCGCAGUUUGCUCUU---ACUUUUAACCCAAUCACCAGCAAAACUU----------- ((((((...(((....((((((((((...))))))..))))....)))...))))))((((((...---..................))))))....----------- ( -20.70, z-score = -2.51, R) >droYak2.chr2L 1793839 94 - 22324452 CUGCGAUUCCGAACUAAUGGCAUAAGCCGCUUAUGUACCAUGUACUCGCACUCGCAGUUUGCUCUU---ACUUUUAACCCACUCAGCAGCAAAACUU----------- ((((((...(((....((((((((((...))))))..))))....)))...))))))((((((((.---...............)).))))))....----------- ( -21.69, z-score = -2.19, R) >droAna3.scaffold_12916 413884 98 + 16180835 UAGCGAUUUCGAACUAAUGGCAUAAGCCGCUUACGUUCCAUGUACUCGUACUCGCAGUUUGCUCUUCUCACUUUUAACCCACUCACCAGCAAAAACUU---------- ..((((....(((((((((((....)))).))).))))...(((....)))))))..((((((........................)))))).....---------- ( -17.56, z-score = -1.61, R) >dp4.chr4_group3 8547208 95 - 11692001 CUGCGAUUCCGAACUAAUGGCAUAAGCCGCUUAUGUACCAUGUACUCGUCCUGGUAGUUUGCUCUU---ACUUUUAACCCACUCACUAGCAAAAACUU---------- ((((.(...(((....((((((((((...))))))..))))....)))...).))))((((((...---..................)))))).....---------- ( -14.80, z-score = -0.02, R) >droPer1.super_1 9999453 95 - 10282868 CUGCGAUUCCGAACUAAUGGCAUAAGCCGCUUAUGUACCAUGUACUCGUCCUGGUAGUUUGCUCUU---ACUUUUAACCCACUCUCUAGCAAAAACUU---------- ((((.(...(((....((((((((((...))))))..))))....)))...).))))((((((...---..................)))))).....---------- ( -14.80, z-score = -0.16, R) >droWil1.scaffold_180708 11853826 105 - 12563649 CUGUGAUGUCGAACUAAUGGCAUAAGCCGCUUAUGCAGUAUGUAUUCCUUGUCGUAGUUGGCUCUU---ACUUUUCGCCCACUGGCCGGAAAAAAAACUUUUUUUUUU ..((((.(((.(((((..(((....)))((....))..................))))))))..))---))((((((((....)).))))))................ ( -19.50, z-score = 0.42, R) >consensus CUGCGAUUCCGAACUAAUGGCAUAAGCCGCUUAUGUACCAUGUACUCGCACUCGCAGUUUGCUCUU___ACUUUUAACCCACUCACCAGCAAAAAUU___________ ((((((...(((....((((((((((...))))))..))))....)))...))))))((((((........................))))))............... (-15.02 = -15.35 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:21 2011