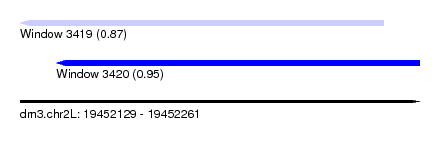
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,452,129 – 19,452,261 |
| Length | 132 |
| Max. P | 0.951708 |

| Location | 19,452,129 – 19,452,249 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Shannon entropy | 0.53382 |
| G+C content | 0.49056 |
| Mean single sequence MFE | -40.51 |
| Consensus MFE | -15.83 |
| Energy contribution | -14.67 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.78 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
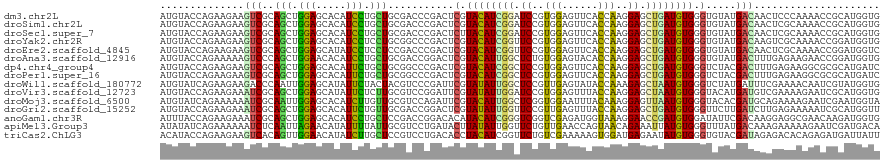
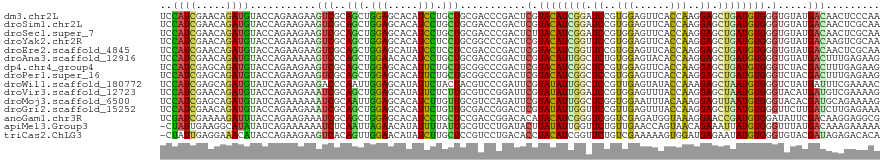
>dm3.chr2L 19452129 120 - 23011544 AUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGAUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCCCAAAACCGCAUGGUG ...(((((......((((((((.(((.....))).)))))))).......(((((((((.(((.(((......))).))).)))))))(((.((......)).)))......)).))))) ( -46.70, z-score = -2.84, R) >droSim1.chr2L 19134319 120 - 22036055 AUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGAUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCGCAAAACCGCAUGGUG ...(((((...((.((((((((.(((.....))).))))))))...((.(.((((((((.(((.(((......))).))).)))))))).).))........))((......)).))))) ( -46.70, z-score = -2.60, R) >droSec1.super_7 3067597 120 - 3727775 AUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCUUACAUCGGAUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCGCAAAACCGCAUGGUG ...(((((...((.((((((((.(((.....))).))))))))...((.((((((((((.(((.(((......))).))).)))))))))).))........))((......)).))))) ( -48.30, z-score = -3.32, R) >droYak2.chr2R 5949462 120 - 21139217 AUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUCCUGCGGCCCGACUCGUACAUCGGUUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAAGUCGCAAAACCGGAUGGUG ...(((((......(((((((..(((.....)))..)))))))(((((.(.((((((((((((.(((......))).)))))))))))).).))..((....)).......))).))))) ( -49.30, z-score = -2.91, R) >droEre2.scaffold_4845 17744432 120 + 22589142 AUGUACCAGAAGAAGUCGCAGCUGGAGCAUAUCCUCCUCCGACCCGACUCGUACAUCGGUUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCGCAAAACCGGAUGGUC ....((((......((((.((..(((.....)))..)).))))(((((.(.((((((((((((.(((......))).)))))))))))).).))..((....)).......))).)))). ( -41.10, z-score = -1.30, R) >droAna3.scaffold_12916 4054542 120 + 16180835 AUGUACCAGAAAAAGUCCCAGCUGGAACACAUCCUGCUGCGACCGGACUCGUACAUUGGCUCUGUGGAGUACACCAAGGAGCUGAUGUGGGUGUAUGACUUUGAGAAGAACCGGAUGGUG ...(((((.......((.((((.(((.....))).)))).))((((((.(.(((((..(((((.(((......))).)))))..))))).).))....(((....)))..)))).))))) ( -44.50, z-score = -2.91, R) >dp4.chr4_group4 552667 120 + 6586962 AUGUACCAGAAGAAGUCGCAGCUGGAGCACAUUCUGCUGCGGCCCGACUCGUACAUCGGCUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUCUACGACUUUGAGAAGGCGCGCAUGAUC ..(..((....(((((((..((((.((((.....)))).))))..(((((..(((((((((((.(((......))).))))))))))))))))..))))))).....))..)........ ( -52.00, z-score = -3.19, R) >droPer1.super_16 859615 120 - 1990440 AUGUACCAGAAGAAGUCGCAGCUGGAGCACAUUCUGCUGCGGCCCGACUCGUACAUCGGCUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUCUACGACUUUGAGAAGGCGCGCAUGAUC ..(..((....(((((((..((((.((((.....)))).))))..(((((..(((((((((((.(((......))).))))))))))))))))..))))))).....))..)........ ( -52.00, z-score = -3.19, R) >droWil1.scaffold_180772 7171607 120 - 8906247 AUGUAUCAGAAGAAGACCCAAUUGGAGCAUAUUCUACUACGUCCCGAUUCGUAUAUUGGCUCCGUUGAGUAUACCAAAGAGCUAAUGUGGGUCUAUGAUUUCGAAAACAAUCGUAUGGUG ...(((((.....((((((((((((..(.((......)).)..))))))...((((((((((..(((.(....)))).))))))))))))))))((((((........)))))).))))) ( -31.90, z-score = -1.80, R) >droVir3.scaffold_12723 632236 120 - 5802038 AUGUACCAGAAGAAAUCGCAGCUGGAGCAUAUUCUCUUGCGUCCGGAUUCGUAUAUUGGAUCCGUGGAGUUUACCAAGGAGCUAAUGUGGGUACAUGAUGUCGAAAAGAAUCGCAUGGUG (((((((.........((...((((((((........))).)))))...))((((((((.(((.(((......))).))).))))))))))))))).(((.(((......)))))).... ( -36.90, z-score = -1.73, R) >droMoj3.scaffold_6500 29143285 120 - 32352404 AUGUAUCAGAAAAAAUCGCAAUUGGAGCACAUCUUGUUGCGUCCAGAUUCGUACAUUGGCUCGGUGGAAUUUACAAAGGAGUUAAUGUGGGUACACGAUGCAGAAAAGAAUCGAAUGGUA .((((((.(((.....((((((.(((.....))).))))))......))).(((((((((((..((.......))...)))))))))))))))))((((.(......).))))....... ( -32.00, z-score = -1.76, R) >droGri2.scaffold_15252 4179294 120 - 17193109 AUGUACCAGAAGAAAUCGCAGCUGGAGCACAUUCUGUUGCGACCGGACUCGUAUAUUGGUUCCGUUGAGUUUACCAAGGAGCUGAUGUGGGUUCUUGAUCUUGAGAAAAAUCGCAUGGUU ....((((...((..(((((((.(((.....))).)))))))..((((((..((((..(((((.(((.(....)))))))))..))))))))))................))...)))). ( -36.60, z-score = -1.42, R) >anoGam1.chr3R 21133030 120 - 53272125 AUUUACCAGAAGAAAUCGCAGCUGGAGCACAUCCUGCUCCGACCGGACACAUACAUCGGGUCGGUCGAGAUGGUAAAGGAACCGAUGUGGAUAUUCGACAAGGAGGCGAACAAGAUGGUG ...(((((.......((((..((((((((.....))))))((((.((........)).)))).((((((.((((......))))((....)).))))))..))..))))......))))) ( -36.12, z-score = -1.26, R) >apiMel3.Group3 12312761 120 + 12341916 AUAUAUCAGAAAAAAUCUCAAUUAGAACAUAUUUUAUUGCGUCCUGAUACUUAUAUUGGUUCUGUUGAACCAGUAACAGAAAUUAUGUGGGUUUAUGACAAAGAAAAAGAAUCGAUGACA ...((((........(((.....)))..............(((.(((.((((((((...((((((((......))))))))...))))))))))).)))..............))))... ( -21.10, z-score = -0.49, R) >triCas2.ChLG3 12606681 120 + 32080666 ACAUACCAGAAGAAGUCACAGUUGGAACAUAUCUUGCUCCGUCCUGACACCUACAUCGGUUCUGUCGAAAAAGUGGAUGAGAAUAUGUGGGUGUACGAUAGAGACACAGAGAUGAUUAUU .....((((..(......)..))))..(((.((((((((..((...((((((((((..((((((((.(.....).))).)))))))))))))))..))..))).)).))).)))...... ( -32.40, z-score = -1.10, R) >consensus AUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGCUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACGAGAAAAACCGCAUGGUG ..............(((..(((.(((.....))).)))...........(.((((((((.((..(((......)))..)).)))))))).).....)))..................... (-15.83 = -14.67 + -1.16)
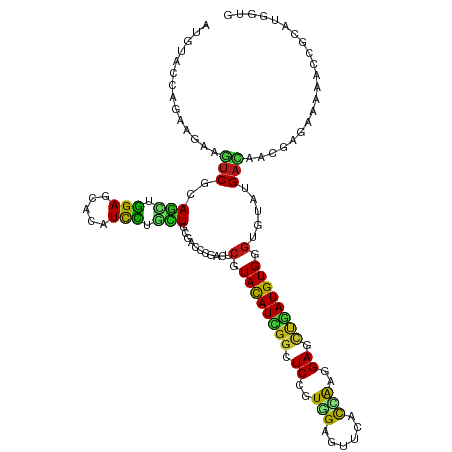
| Location | 19,452,141 – 19,452,261 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.28 |
| Shannon entropy | 0.49209 |
| G+C content | 0.49041 |
| Mean single sequence MFE | -40.58 |
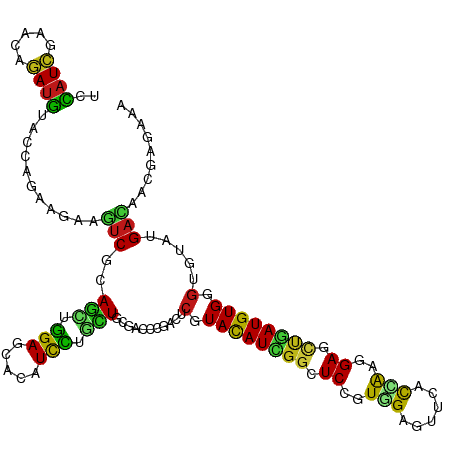
| Consensus MFE | -18.31 |
| Energy contribution | -16.83 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.78 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19452141 120 - 23011544 UCCAUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGAUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCCCAA ..((((.....(((((((.((..(..((((((((.(((.....))).)))))))).)..)).)))))))((.(((.(((......))).))).))))))((((.((......)).)))). ( -48.50, z-score = -3.62, R) >droSim1.chr2L 19134331 120 - 22036055 UCCAUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGAUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCGCAA (((((.((.(.(((((((.((..(..((((((((.(((.....))).)))))))).)..)).))))))).).)).)))))((((......))))...(((((((........))))))). ( -49.40, z-score = -3.61, R) >droSec1.super_7 3067609 120 - 3727775 UCCAUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCUUACAUCGGAUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCGCAA (((((.((.(.((((((..((..(..((((((((.(((.....))).)))))))).)..))..)))))).).)).)))))((((......))))...(((((((........))))))). ( -45.80, z-score = -2.84, R) >droYak2.chr2R 5949474 120 - 21139217 UCCAUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUCCUGCGGCCCGACUCGUACAUCGGUUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAAGUCGCAA ..((((.....))))...........(((((((..(((.....)))..))))))).((((((.((((((((((((.(((......))).)))))))))))).)((.....)))))))... ( -45.80, z-score = -2.32, R) >droEre2.scaffold_4845 17744444 120 + 22589142 UCCAUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCAUAUCCUCCUCCGACCCGACUCGUACAUCGGUUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCGCAA (((((.((((.(((((((.((..(..((((.((..(((.....)))..)).)))).)..)).))))))).)))).)))))((((......))))...(((((((........))))))). ( -43.70, z-score = -2.50, R) >droAna3.scaffold_12916 4054554 120 + 16180835 UCCAUCGAACAGAUGUACCAGAAAAAGUCCCAGCUGGAACACAUCCUGCUGCGACCGGACUCGUACAUUGGCUCUGUGGAGUACACCAAGGAGCUGAUGUGGGUGUAUGACUUUGAGAAG ..((((.....))))...((((....(((.((((.(((.....))).)))).)))((.((.(.(((((..(((((.(((......))).)))))..))))).).)).))..))))..... ( -40.90, z-score = -2.18, R) >dp4.chr4_group4 552679 120 + 6586962 UCCAUCGAGCAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUUCUGCUGCGGCCCGACUCGUACAUCGGCUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUCUACGACUUUGAGAAG ..((((.....))))........(((((((..((((.((((.....)))).))))..(((((..(((((((((((.(((......))).))))))))))))))))..)))))))...... ( -51.20, z-score = -3.37, R) >droPer1.super_16 859627 120 - 1990440 UCCAUCGAGCAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUUCUGCUGCGGCCCGACUCGUACAUCGGCUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUCUACGACUUUGAGAAG ..((((.....))))........(((((((..((((.((((.....)))).))))..(((((..(((((((((((.(((......))).))))))))))))))))..)))))))...... ( -51.20, z-score = -3.37, R) >droWil1.scaffold_180772 7171619 120 - 8906247 UCCAUCGAGCAGAUGUAUCAGAAGAAGACCCAAUUGGAGCAUAUUCUACUACGUCCCGAUUCGUAUAUUGGCUCCGUUGAGUAUACCAAAGAGCUAAUGUGGGUCUAUGAUUUCGAAAAC ..((((.....)))).((((.....((((((((((((..(.((......)).)..))))))...((((((((((..(((.(....)))).)))))))))))))))).))))......... ( -31.80, z-score = -1.85, R) >droVir3.scaffold_12723 632248 120 - 5802038 UCCAUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCAUAUUCUCUUGCGUCCGGAUUCGUAUAUUGGAUCCGUGGAGUUUACCAAGGAGCUAAUGUGGGUACAUGAUGUCGAAAAG ....((((.((.(((((((.........((...((((((((........))).)))))...))((((((((.(((.(((......))).))).)))))))))))))))..)))))).... ( -39.60, z-score = -2.80, R) >droMoj3.scaffold_6500 29143297 120 - 32352404 UCCAUCGAGCAGAUGUAUCAGAAAAAAUCGCAAUUGGAGCACAUCUUGUUGCGUCCAGAUUCGUACAUUGGCUCGGUGGAAUUUACAAAGGAGUUAAUGUGGGUACACGAUGCAGAAAAG ((((((((((.(((((((..(((.....((((((.(((.....))).))))))......)))))))))).))))))))))............((...((((....))))..))....... ( -38.90, z-score = -3.58, R) >droGri2.scaffold_15252 4179306 120 - 17193109 UCCAUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCACAUUCUGUUGCGACCGGACUCGUAUAUUGGUUCCGUUGAGUUUACCAAGGAGCUGAUGUGGGUUCUUGAUCUUGAGAAA (((.((((((((((((.((((..(....)....)))).)))...)))))).)))..)))..(.(((((..(((((.(((.(....)))))))))..))))).)(((((......))))). ( -36.90, z-score = -1.47, R) >anoGam1.chr3R 21133042 120 - 53272125 UCGAUCGAAAAGAUUUACCAGAAGAAAUCGCAGCUGGAGCACAUCCUGCUCCGACCGGACACAUACAUCGGGUCGGUCGAGAUGGUAAAGGAACCGAUGUGGAUAUUCGACAAGGAGGCG ..(.(((((........((((..(....)....))))..((((((((...((((((.((........)).))))))...))..(((......)))))))))....))))))......... ( -32.90, z-score = -0.23, R) >apiMel3.Group3 12312773 119 + 12341916 -CUAUUGAAGGCAUAUAUCAGAAAAAAUCUCAAUUAGAACAUAUUUUAUUGCGUCCUGAUACUUAUAUUGGUUCUGUUGAACCAGUAACAGAAAUUAUGUGGGUUUAUGACAAAGAAAAA -.........(((......(((.....)))....(((((....))))).)))(((.(((.((((((((...((((((((......))))))))...))))))))))).)))......... ( -20.00, z-score = -0.18, R) >triCas2.ChLG3 12606693 119 + 32080666 -CUAUUGAGGAAACAUACCAGAAGAAGUCACAGUUGGAACAUAUCUUGCUCCGUCCUGACACCUACAUCGGUUCUGUCGAAAAAGUGGAUGAGAAUAUGUGGGUGUACGAUAGAGACACA -((((((.(....)................(((.((((.((.....)).))))..)))((((((((((..((((((((.(.....).))).))))))))))))))).))))))....... ( -32.10, z-score = -0.93, R) >consensus UCCAUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGCUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACGAGAAA ..((((.....))))...........(((..(((.(((.....))).)))...........(.((((((((.((..(((......)))..)).)))))))).).....)))......... (-18.31 = -16.83 + -1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:08 2011