| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,441,389 – 19,441,449 |
| Length | 60 |
| Max. P | 0.551637 |

| Location | 19,441,389 – 19,441,449 |
|---|---|
| Length | 60 |
| Sequences | 9 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.62418 |
| G+C content | 0.44742 |
| Mean single sequence MFE | -12.28 |
| Consensus MFE | -5.03 |
| Energy contribution | -4.46 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.89 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19441389 60 + 23011544 -GGUCGGUCGGAAUAGUCGGGCACUCGUAAGGCCAGCGCGACUUAAUAGGUCAUUAUAAGC--- -....(..(.....(((((.((..((....))...)).))))).....)..).........--- ( -16.00, z-score = -1.27, R) >droPer1.super_16 843961 55 + 1990440 AGUCCAGUCAGAAUAGUCUGUCA--UAUUAAGCCUC----AUUUAAUAGGCCAUUAAAAGU--- ......(.((((....)))).).--......((((.----.......))))..........--- ( -8.10, z-score = -2.33, R) >dp4.chr4_group4 537828 55 - 6586962 AGUCCAGUCAGAAUAGUCUGUCA--UAUUAAGCCAC----AUUUAAUAGGCCAUUAAAAGU--- .(.((.(.((((....)))).).--(((((((....----.))))))))).).........--- ( -7.20, z-score = -1.95, R) >droAna3.scaffold_12916 1303946 58 - 16180835 -CGUCAACCAGAAUAGUCGGGUU--UAUAAGGCCAAGACGACUUAAUAGGUCAUUAUAAGC--- -..............(((.((((--.....))))..)))((((.....)))).........--- ( -9.80, z-score = -1.05, R) >droEre2.scaffold_4845 17733859 60 - 22589142 -UGUCGGUCGGAAAAGUCGGGCACUCGUAAGGCCAACGCGACUUAAUAGGUCAUUAUAAGC--- -....(..(....(((((((((.((....)))))....))))))....)..).........--- ( -13.50, z-score = -0.65, R) >droYak2.chr2R 5938729 60 + 21139217 -UGUCGGUCGGAAUAGUCGGGCACUCGUAAGGCCAACGCGACUUAAUAGGUCAUUAUAAGC--- -....(..(.....((((((((.((....)))))....))))).....)..).........--- ( -13.10, z-score = -0.33, R) >droSec1.super_7 3056811 60 + 3727775 -GGUCGGUCGGAAUAGUCGGGCACUCGUAAGACCAGCGCGACUUAAUAGGUCAUUAUAAGC--- -....(..(.....(((((.((..((....))...)).))))).....)..).........--- ( -16.90, z-score = -1.80, R) >droSim1.chr2L 19122932 60 + 22036055 -GGUCGGUCGGAAUAGUCGGGCACUCGUAAGGCCAGCGCGACUUAAUAGGUCAUUAUAAGC--- -....(..(.....(((((.((..((....))...)).))))).....)..).........--- ( -16.00, z-score = -1.27, R) >droVir3.scaffold_12723 3366181 62 + 5802038 -UGUAUGUGUGCACACACAAACACAUCGAAAAGUAAAAC-ACAAAGUGAUUUAUUAUGGCUAGU -(((.(((((....))))).)))........((((((.(-((...))).))))))......... ( -9.90, z-score = 0.09, R) >consensus _GGUCGGUCGGAAUAGUCGGGCACUCGUAAGGCCAACGCGACUUAAUAGGUCAUUAUAAGC___ ......(((.((....)).)))........((((..............))))............ ( -5.03 = -4.46 + -0.56)

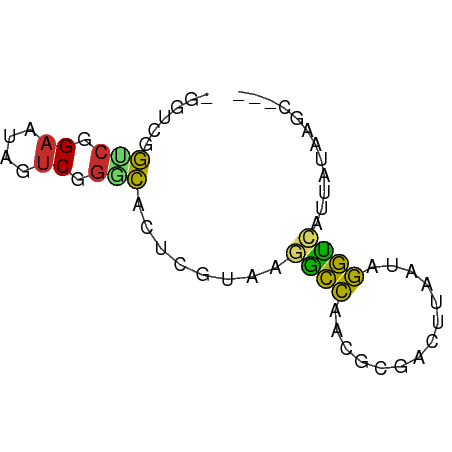
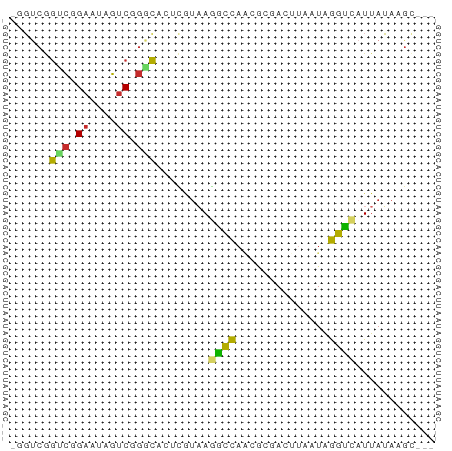
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:06 2011