
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,412,909 – 19,413,003 |
| Length | 94 |
| Max. P | 0.982547 |

| Location | 19,412,909 – 19,413,003 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Shannon entropy | 0.40555 |
| G+C content | 0.37538 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -12.75 |
| Energy contribution | -13.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19412909 94 + 23011544 UCGAGUGAAAACUCAGCCACAGUCUGCCUGCCUACCGCUCAAAUUAUGAGCUUUUCUUUUCUAUAUAUGUAUGUAUGAACA-AUAUAUGAAAAGG ..((((....)))).....(((.....)))......(((((.....)))))....((((((.((((((((........)).-)))))))))))). ( -20.90, z-score = -2.41, R) >droSim1.chr2L 19094429 88 + 22036055 UCGAGUGGAAACUCAGCCACAGUGUGCCU-UCUAUCGCUCAAAUUAUGAGCUUUUCUUUUCUACACAUAUGUACAU------AUAUAUGAAAAGG ..((((....))))............(((-(.....(((((.....)))))..............(((((((....------)))))))..)))) ( -18.50, z-score = -1.57, R) >droSec1.super_7 3028726 88 + 3727775 UCGAGUGGAAACUCAGCCACAGUGUGCCU-UCUAUCGCUCAAAUUGUGAGCUUUUCUUUUCUAUACAUAUGUACAU------AUAUAUGAAGAGG ..((((....))))(((((((((..((..-......))....)))))).)))...((((((.(((.((((....))------)).))))))))). ( -22.90, z-score = -2.25, R) >droEre2.scaffold_4845 17705917 87 - 22589142 UCGAGUGGAAACUCGACCAC----UGCAA----ACCACUCAAAUUAUGAGUUUUAUGUACAUACAUAUGUACAUGUGCAUACAUCUGUAUAAAGG ((((((....))))))((..----.(((.----...(((((.....)))))..(((((((((....))))))))))))((((....))))...)) ( -26.00, z-score = -3.90, R) >consensus UCGAGUGGAAACUCAGCCACAGUGUGCCU_UCUACCGCUCAAAUUAUGAGCUUUUCUUUUCUACACAUAUAUACAU______AUAUAUGAAAAGG ..((((....))))..((..................(((((.....)))))..............(((((((..........)))))))....)) (-12.75 = -13.25 + 0.50)

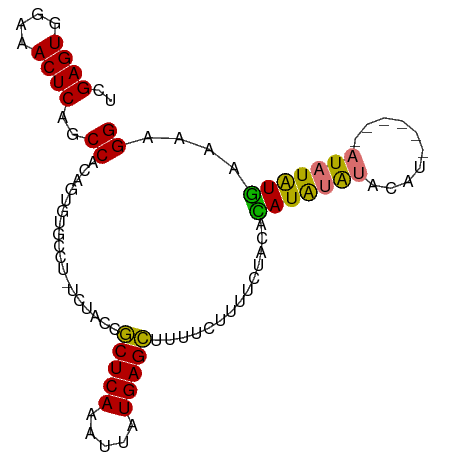
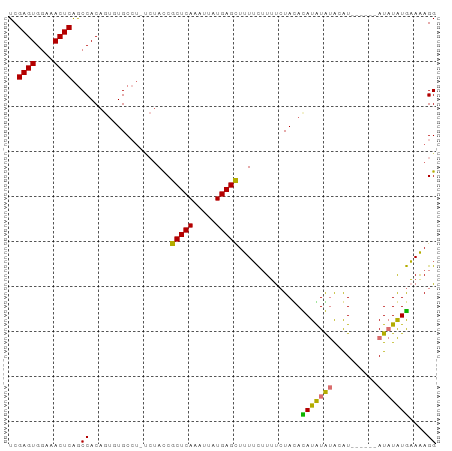
| Location | 19,412,909 – 19,413,003 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Shannon entropy | 0.40555 |
| G+C content | 0.37538 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -12.29 |
| Energy contribution | -13.17 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19412909 94 - 23011544 CCUUUUCAUAUAU-UGUUCAUACAUACAUAUAUAGAAAAGAAAAGCUCAUAAUUUGAGCGGUAGGCAGGCAGACUGUGGCUGAGUUUUCACUCGA .((((((((((((-.((........)))))))).))))))....(((((.....)))))(((..((((.....)))).)))((((....)))).. ( -25.10, z-score = -2.59, R) >droSim1.chr2L 19094429 88 - 22036055 CCUUUUCAUAUAU------AUGUACAUAUGUGUAGAAAAGAAAAGCUCAUAAUUUGAGCGAUAGA-AGGCACACUGUGGCUGAGUUUCCACUCGA .((((((((((((------((....)))))))).))))))....(((((.....)))))......-((.(((...))).))((((....)))).. ( -24.40, z-score = -2.83, R) >droSec1.super_7 3028726 88 - 3727775 CCUCUUCAUAUAU------AUGUACAUAUGUAUAGAAAAGAAAAGCUCACAAUUUGAGCGAUAGA-AGGCACACUGUGGCUGAGUUUCCACUCGA .((.(((((((((------((....)))))))).))).))....(((((.....)))))......-((.(((...))).))((((....)))).. ( -20.00, z-score = -1.34, R) >droEre2.scaffold_4845 17705917 87 + 22589142 CCUUUAUACAGAUGUAUGCACAUGUACAUAUGUAUGUACAUAAAACUCAUAAUUUGAGUGGU----UUGCA----GUGGUCGAGUUUCCACUCGA ....(((((....)))))(((((((((((....))))))))...(((((.....)))))...----.....----))).((((((....)))))) ( -23.10, z-score = -1.75, R) >consensus CCUUUUCAUAUAU______AUAUACACAUAUAUAGAAAAGAAAAGCUCAUAAUUUGAGCGAUAGA_AGGCACACUGUGGCUGAGUUUCCACUCGA .((((((((((((..............)))))).))))))....(((((.....)))))........(((........)))((((....)))).. (-12.29 = -13.17 + 0.88)

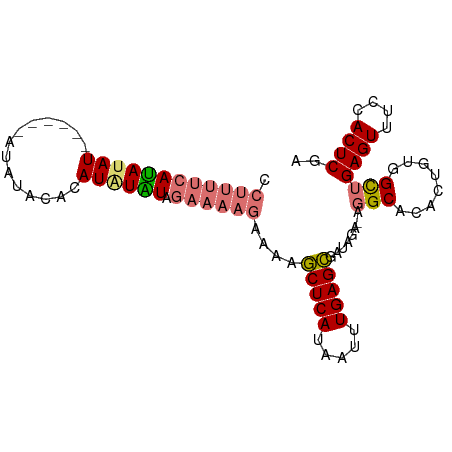
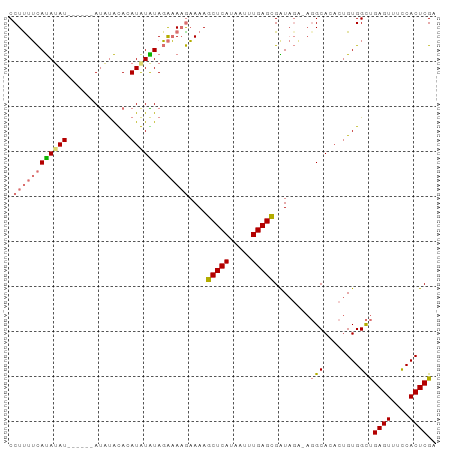
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:05 2011