
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,396,622 – 19,396,736 |
| Length | 114 |
| Max. P | 0.580076 |

| Location | 19,396,622 – 19,396,736 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 68.33 |
| Shannon entropy | 0.59984 |
| G+C content | 0.38378 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -8.73 |
| Energy contribution | -8.43 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.578827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
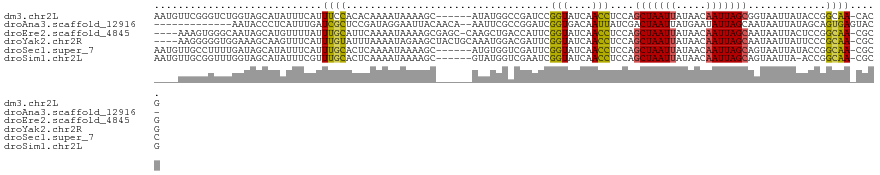
>dm3.chr2L 19396622 114 + 23011544 AAUGUUCGGGUCUGGUAGCAUAUUUCAUUUCCACACAAAAUAAAAGC------AUAUGGCCGAUCCGGUAUCAACCUCCAGCUAAUUAUAACAAUUAGCGGUAAUUAUACCGGCAA-CACG .......(((((.(((..(((((........................------)))))))))))))(((((......((.(((((((.....))))))))).....)))))(....-)... ( -24.36, z-score = -1.43, R) >droAna3.scaffold_12916 11153481 105 - 16180835 -------------AAUACCCUCAUUUGAUCGCUCCGAUAGGAAUUACAACA--AAUUCGCCGGAUCGGUGACAAUUAUCGACUAAUUAUGAAUAUUAGCAAUAAUUAUAGCAGUGAGUAC- -------------......((((((..((((.((((....(((((......--)))))..)))).))))...((((((.(.(((((.......)))))).)))))).....))))))...- ( -19.70, z-score = -0.83, R) >droEre2.scaffold_4845 17691143 115 - 22589142 ----AAAGUGGGCAAUAGCAUGUUUUAUUUGCAUUCAAAAUAAAAGCGAGC-CAAGCUGACCAUUCGGUAUCAACCUCCAGCUAAUUAUAACAAUUAGCAAUAAUUACUCCGGCAA-CGCG ----...((((((....((...((((((((.......))))))))))..))-)..(((((((....)))...........(((((((.....)))))))...........))))..-))). ( -25.50, z-score = -1.68, R) >droYak2.chr2R 5894786 116 + 21139217 ----AAGGGGGUGGAAAGCAAGUUUCAUUUGUAUUUAAAAUAGAAGCUACUGCAAAUGGACGAUUCGGUAUCAACCUCCAGCUAAUUAUAACAAUUAGCAAUAAUUAUUCCCGCAA-CGCG ----..(((((((((..(((((((((((((.......)))).))))))..)))....((..(((.....)))..))))).(((((((.....))))))).......))))))....-.... ( -28.60, z-score = -2.26, R) >droSec1.super_7 3012891 114 + 3727775 AAUGUUGCCUUUUGAUAGCAUAUUUCAUUUGCACUCAAAAUAAAAGC------AUGUGGUCGAUUCGGUAUCAACCUCCAGCUAAUUAUAACAAUUAGCAGUAAUUAUACCGGCAA-CGCC ...((((((..(((((.((((.(((.((((.......)))).)))..------)))).)))))...(((((.........(((((((.....))))))).......))))))))))-)... ( -27.29, z-score = -2.65, R) >droSim1.chr2L 19078305 113 + 22036055 AAUGUUGCGGUUUGGUAGCAUAUUUCGUUUGCACUCAAAAUAAAAGC------GUAUGGUCGAAUCGGUAUCAACCUCCAGCUAAUUAUAACAAUUAGCAGUAAUUA-ACCGGCAA-CGCG .(((((((......)))))))........................((------((...((((....(((....)))....(((((((.....)))))))........-..)))).)-))). ( -25.50, z-score = -0.97, R) >consensus ____UUGGGGUUUGAUAGCAUAUUUCAUUUGCACUCAAAAUAAAAGC______AAAUGGACGAUUCGGUAUCAACCUCCAGCUAAUUAUAACAAUUAGCAAUAAUUAUACCGGCAA_CGCG ............................((((..................................(((....)))....(((((((.....))))))).............))))..... ( -8.73 = -8.43 + -0.30)

| Location | 19,396,622 – 19,396,736 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 68.33 |
| Shannon entropy | 0.59984 |
| G+C content | 0.38378 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -9.73 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
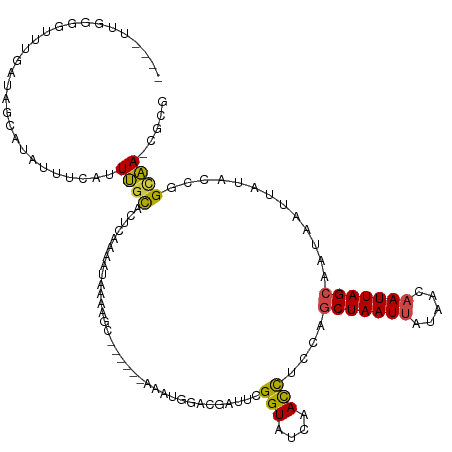
>dm3.chr2L 19396622 114 - 23011544 CGUG-UUGCCGGUAUAAUUACCGCUAAUUGUUAUAAUUAGCUGGAGGUUGAUACCGGAUCGGCCAUAU------GCUUUUAUUUUGUGUGGAAAUGAAAUAUGCUACCAGACCCGAACAUU .(((-(..((((((((((..((((((((((...)))))))).))..))).))))))).((((......------((...(((((..(......)..))))).))........)))))))). ( -31.34, z-score = -1.91, R) >droAna3.scaffold_12916 11153481 105 + 16180835 -GUACUCACUGCUAUAAUUAUUGCUAAUAUUCAUAAUUAGUCGAUAAUUGUCACCGAUCCGGCGAAUU--UGUUGUAAUUCCUAUCGGAGCGAUCAAAUGAGGGUAUU------------- -((((((..(((((((((((((((((((.......))))).))))))))))..(((((..((.(((((--......)))))))))))))))).((....)))))))).------------- ( -27.00, z-score = -2.28, R) >droEre2.scaffold_4845 17691143 115 + 22589142 CGCG-UUGCCGGAGUAAUUAUUGCUAAUUGUUAUAAUUAGCUGGAGGUUGAUACCGAAUGGUCAGCUUG-GCUCGCUUUUAUUUUGAAUGCAAAUAAAACAUGCUAUUGCCCACUUU---- .(((-..(((............((((((((...))))))))...((((((((........)))))))))-)).)))((((((((.......))))))))...((....)).......---- ( -24.70, z-score = 0.45, R) >droYak2.chr2R 5894786 116 - 21139217 CGCG-UUGCGGGAAUAAUUAUUGCUAAUUGUUAUAAUUAGCUGGAGGUUGAUACCGAAUCGUCCAUUUGCAGUAGCUUCUAUUUUAAAUACAAAUGAAACUUGCUUUCCACCCCCUU---- ....-....(((..........((((((((...))))))))((((((.((((.....)))).))....(((..((.(((.((((.......))))))).)))))..))))..)))..---- ( -20.90, z-score = 0.27, R) >droSec1.super_7 3012891 114 - 3727775 GGCG-UUGCCGGUAUAAUUACUGCUAAUUGUUAUAAUUAGCUGGAGGUUGAUACCGAAUCGACCACAU------GCUUUUAUUUUGAGUGCAAAUGAAAUAUGCUAUCAAAAGGCAACAUU ...(-((((((((((.....(.((((((((...)))))))).)..(((((((.....)))))))..))------)))....((((((..(((.........)))..))))))))))))... ( -33.50, z-score = -3.20, R) >droSim1.chr2L 19078305 113 - 22036055 CGCG-UUGCCGGU-UAAUUACUGCUAAUUGUUAUAAUUAGCUGGAGGUUGAUACCGAUUCGACCAUAC------GCUUUUAUUUUGAGUGCAAACGAAAUAUGCUACCAAACCGCAACAUU ...(-((((.(((-(.......((((((((...))))))))(((.((((((.......))))))....------((...(((((((........))))))).))..))))))))))))... ( -33.00, z-score = -3.53, R) >consensus CGCG_UUGCCGGUAUAAUUACUGCUAAUUGUUAUAAUUAGCUGGAGGUUGAUACCGAAUCGACCAUAU______GCUUUUAUUUUGAGUGCAAAUGAAAUAUGCUAUCAAACCCCAA____ .........((((((((((.(((((((((.....))))))).)).)))).))))))..................((...(((((((........))))))).))................. ( -9.73 = -10.35 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:01 2011