
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,385,142 – 19,385,250 |
| Length | 108 |
| Max. P | 0.732041 |

| Location | 19,385,142 – 19,385,250 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.61 |
| Shannon entropy | 0.52674 |
| G+C content | 0.61771 |
| Mean single sequence MFE | -43.69 |
| Consensus MFE | -22.29 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19385142 108 - 23011544 ------AUGACUCUGCUCUGGGAAAACUCGAGAGCUCCUACGACUCCGUGGGUGCGACUGAAUCCGCCGAAGGUGCC---UCCGCUGGCGGAUUAGGUGGUGCCGUUGAGACUUUGG ------....(((.((((((((....))).)))))...((((....))))((..(.(((.(((((((((..((....---.))..))))))))).))).)..))...)))....... ( -42.80, z-score = -2.12, R) >droSim1.chr2L 19066829 108 - 22036055 ------AAGGCUCCUCUCUGGGAAAACUGGAGAGCUCCUACGACUCCGUGGGUGCGACUGAAUCCGCCGAAGGUGCC---UCCGCUGGCGGAUUAGGUGGUGCCGUUGAGACUUUGG ------..(((.((((((..(.....)..))))((.((((((....)))))).)).(((.(((((((((..((....---.))..))))))))).))))).)))............. ( -48.90, z-score = -3.05, R) >droSec1.super_7 3001436 108 - 3727775 ------AAGGCUCCUCUCUGGGAAAACUGGAGAGCUCCUACGACUCCGUGGGUGCGACUGAAUCCGCCGAAGGUGCC---UCCGCUGGCGGAUUAGGUGGUGCCGUUGAGACUUUGG ------..(((.((((((..(.....)..))))((.((((((....)))))).)).(((.(((((((((..((....---.))..))))))))).))))).)))............. ( -48.90, z-score = -3.05, R) >droYak2.chr2R 5882930 105 - 21139217 ------AUGGCGCAGCGUUGGGAAACCUGGAGAGCUCCUACGACUCCGUGGGUGCCACUGAAUCCGCUGAGGGUGCC---UCCGCUGGAGG---UGCUGGUGGCGUUGAGCUUUUGG ------..(((.((((...((....)).(((..((.((((((....)))))).)).......)))((((..((..((---(((...)))))---..))..)))))))).)))..... ( -44.10, z-score = -1.05, R) >droEre2.scaffold_4845 17679341 108 + 22589142 ------AUGGCUCGGCUUUGGGAAACCUGGAGAGCUCCUACGACUCCGUGGGUGCCACUGAGUCCGUCGAGGGUGCC---UCCGCUGGAGGAUUAGGUGGUGCCGUGGAGACUUUGG ------..(((((..((..((....)).)).))))).......(((((((.(..(((((.(((((.((.((((....---.)).)).))))))).)))))..))))))))....... ( -46.10, z-score = -1.29, R) >droAna3.scaffold_12916 11141499 108 + 16180835 ACUCCUCUGCCGGAAACUUGGGCAACUUGGAGAGCUCCUAUGAGUCUGUGGGUGUGACAGAGUCCGCCGCCGGUGCCGGAUCAGCUGGAAGCUCUGUGGAUUCCAUUG--------- .((((..(((((....)...))))....)))).((((....))))..(((((..(.(((((((((((..(((....)))....)).))..))))))).)..)))))..--------- ( -36.70, z-score = 0.23, R) >dp4.chr4_group4 482226 99 + 6586962 ---------GCGGCAGUCUGGGUAACCUGGAGAGCUCCUACGAUUCCGUGGGCGCCGUAGAGGCCGCUGAAGGCGCC---UCCGGCCAGGGA------AGUGCCAGCGAGACUUUUG ---------(((((..((..((...))..)).....((((((....)))))).)))))((((((((((...(((((.---(((......)))------.))))))))).).))))). ( -41.00, z-score = -0.19, R) >droPer1.super_16 788287 99 - 1990440 ---------GCGGCAGUCUGGGUAACCUGGAGAGCUCCUACGAUUCCGUGGGCGCCGUAGAGGCCGCUGAAGGCGCC---UCCGGCCAGGGA------AGUGCCAGCGAGACUUUUG ---------(((((..((..((...))..)).....((((((....)))))).)))))((((((((((...(((((.---(((......)))------.))))))))).).))))). ( -41.00, z-score = -0.19, R) >consensus ______AUGGCGCCACUCUGGGAAACCUGGAGAGCUCCUACGACUCCGUGGGUGCCACUGAAUCCGCCGAAGGUGCC___UCCGCUGGAGGAUUAGGUGGUGCCGUUGAGACUUUGG ........(((.....((..(.....)..))..((.((((((....)))))).))..........)))...((((((...(((......)))......))))))............. (-22.29 = -23.06 + 0.77)

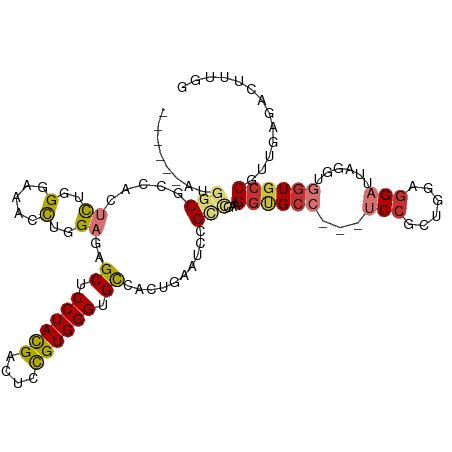
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:59 2011