| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,819,701 – 1,819,807 |
| Length | 106 |
| Max. P | 0.905028 |

| Location | 1,819,701 – 1,819,807 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.41172 |
| G+C content | 0.42176 |
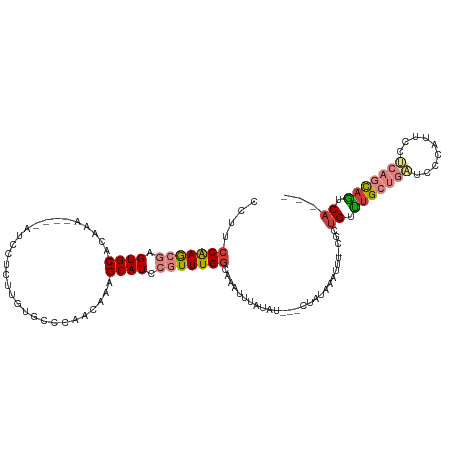
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -12.34 |
| Energy contribution | -14.31 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
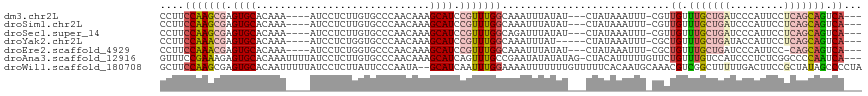
>dm3.chr2L 1819701 106 + 23011544 CCUUCCAAGCGAGUGCACAAA----AUCCUCUUGUGCCCAACAAAGCAUCCGUUUGGCAAAUUUAUAU---CUAUAAAUUU-CGUUGUUUGCUGAUCCCAUUCCUCAGCAGUCA--- ....(((((((..(((((((.----......))))(.....)...)))..))))))).((((((((..---..))))))))-...((.(((((((.........))))))).))--- ( -22.40, z-score = -2.72, R) >droSim1.chr2L 1803844 106 + 22036055 CCUUCCAAGCGAGUGCACAAA----AUCCUCUUGUGCCCAACAAAGCAUCCGUUUGGCAAAUUUAUAU---CUAUAAAUUU-CGUUGUUUGCUGAUCCCAUUCCUCAGCAGUCA--- ....(((((((..(((((((.----......))))(.....)...)))..))))))).((((((((..---..))))))))-...((.(((((((.........))))))).))--- ( -22.40, z-score = -2.72, R) >droSec1.super_14 1766374 106 + 2068291 CCUUCCAAGCGAGUGCACAAA----AUCCUCUUGUGCCCAACAAAGCAUCCGUUUGGCAGAUUUAUAU---CUAUAAAUUU-CGUUGUUUGCUGAUCCCAUUCCUCAGCAGUCA--- .((.(((((((..(((((((.----......))))(.....)...)))..))))))).))((((((..---..))))))..-...((.(((((((.........))))))).))--- ( -22.60, z-score = -2.23, R) >droYak2.chr2L 1792743 104 + 22324452 CCUUCCAAACGAGUGCACAAA----AUCCUCUGGUGCCCAACAAAGCAUCCGUUUGGCAAAUUUAU-----CUAUAAAUUU-CGCUGUUUGCUGAUACCAUUCCUCAGCAGUCA--- .....((((((..(((.((..----......))((.....))...)))..))))))(((((((((.-----...)))))))-.)).(.(((((((.........))))))).).--- ( -22.80, z-score = -2.34, R) >droEre2.scaffold_4929 1864602 105 + 26641161 CCUUCCAAACGAGUGCACAAA----AUCCUCUGGUGCCCAACAAAGCAUCCGUUUGGCAAAUUUAUAU---CUAUAAAUUU-CGCUGUUUGCUGAUCCCAUUCC-CAGCAGUCA--- .....((((((..(((.((..----......))((.....))...)))..))))))((((((((((..---..))))))))-.)).(.((((((..........-)))))).).--- ( -22.80, z-score = -2.48, R) >droAna3.scaffold_12916 412695 113 - 16180835 GUUUCCGAAAGAGUGCACAAAUUUUAUCCUCUUGUGCCCAACAAAGCAUCAGUUUGCCGAAUAUAUAUAG-CUACAUUUUUGUUCUGUUUGUCCAUCCCUCUCGGCCCCAAUCA--- ....((((.((.(.((((((...........)))))).).((((((((......))).(((((.......-.........)))))..)))))......)).)))).........--- ( -17.19, z-score = -0.83, R) >droWil1.scaffold_180708 11852953 115 + 12563649 GCUUCCAAGCGAGUGCACAAUUUUUAUCCUCUUAUUCCCAAUA--GCAUCAAUUUGGAAAAUUUUUUUGUUUUUCACAAUGCAAACGUCGGCUUUUUGACUUCCGCUAUAGCCCCUA (((....((((................................--((((.....((((((((......))))))))..))))....(((((....)))))...))))..)))..... ( -17.70, z-score = -0.52, R) >consensus CCUUCCAAGCGAGUGCACAAA____AUCCUCUUGUGCCCAACAAAGCAUCCGUUUGGCAAAUUUAUAU___CUAUAAAUUU_CGCUGUUUGCUGAUCCCAUUCCUCAGCAGUCA___ ....(((((((.((((.............................)))).))))))).(((((((((.....))))))))).....(.(((((((.........))))))).).... (-12.34 = -14.31 + 1.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:20 2011