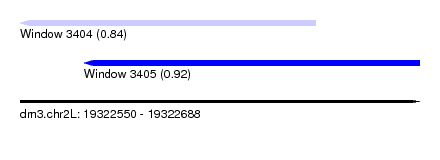
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,322,550 – 19,322,688 |
| Length | 138 |
| Max. P | 0.916453 |

| Location | 19,322,550 – 19,322,652 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Shannon entropy | 0.10803 |
| G+C content | 0.45045 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -25.33 |
| Energy contribution | -25.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
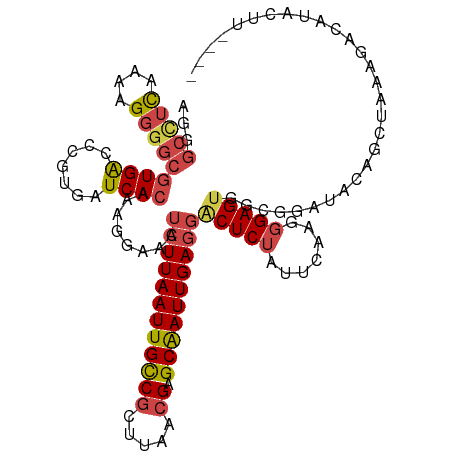
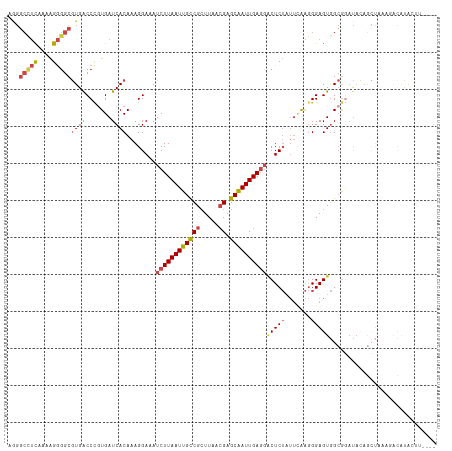
>dm3.chr2L 19322550 102 - 23011544 GAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUCAAGGGAGUGGCGGAUACAGCUAAAGACAUACUUAUAAGGCGGUAGAAGUGCUGCAUACG ....((((((((((((.....)).))))))))))(((((.......)))))(((.......))).................((((((....))))))..... ( -25.30, z-score = -1.10, R) >droSim1.chr2L 19015571 102 - 22036055 GAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUCGAGGGAGUGGCGGAUACAGCUAAAGACAUACUUAUAAGGCGGUAGAAGUGCUGCAAACG ....((((((((((((.....)).))))))))))(((((.......)))))(((.......))).................((((((....))))))..... ( -25.30, z-score = -0.98, R) >droYak2.chr2R 5830175 98 - 21139217 GAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUAGAGGGAGUGGCGGAUAUAGCUGAAGACA----UAUACGGCGGUAAGAGUGCUGCAUACG ....((((((((((((.....)).))))))))))(((((.......)))))(((.......))).......----......((((((....))))))..... ( -25.40, z-score = -1.28, R) >consensus GAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUCGAGGGAGUGGCGGAUACAGCUAAAGACAUACUUAUAAGGCGGUAGAAGUGCUGCAUACG ....((((((((((((.....)).))))))))))(((((.......)))))(((.......))).................((((((....))))))..... (-25.33 = -25.33 + -0.00)

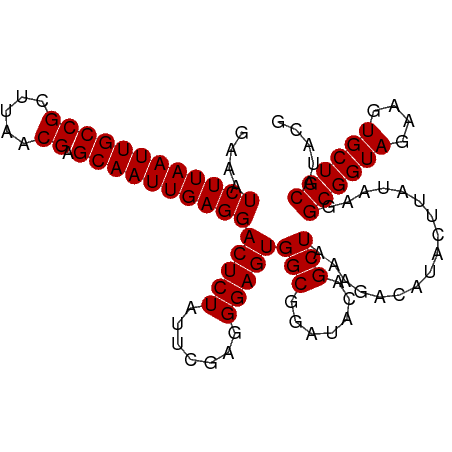
| Location | 19,322,572 – 19,322,688 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.80 |
| Shannon entropy | 0.47333 |
| G+C content | 0.46648 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
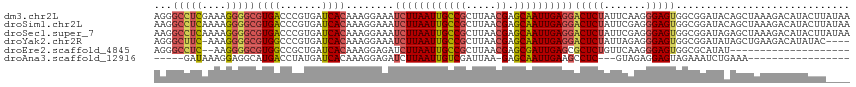
>dm3.chr2L 19322572 116 - 23011544 AGGGCCUCGAAAGGGGCGUGACCCGUGAUCACAAAGGAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUCAAGGGAGUGGCGGAUACAGCUAAAGACAUACUUAUAA ((((((((....)))))(((.(((.(((.......(((..((((((((((((.....)).))))))))))..)))..))).)))..((((.......))))....))).))).... ( -35.60, z-score = -2.36, R) >droSim1.chr2L 19015593 116 - 22036055 AAGGCCUCAAAAGGGGCGUGACCCGUGAUCACAAAGGAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUCGAGGGAGUGGCGGAUACAGCUAAAGACAUACUUAUAA ((((((((....)))))(((..((((.((..(........((((((((((((.....)).)))))))))).(((.....))))..)).)))).))).............))).... ( -32.30, z-score = -1.59, R) >droSec1.super_7 2950833 116 - 3727775 AAGGCCUCAAAAGGGGCGUGACCCGUGAUCACAAAGGAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUCGAGGGAGUGGCGGAUAGAGCUAAAGACAUACUUAUAA ...(((((....)))))((((.......)))).(((....((((((((((((.....)).)))))))))).(((((((((.........)))))))))...........))).... ( -33.80, z-score = -1.90, R) >droYak2.chr2R 5830197 111 - 21139217 AGGGCUUC-AAAGGGGCGUGGCCCGUGAUCACAAAGGAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUAGAGGGAGUGGCGGAUAUAGCUGAAGACAUAUAC---- .(..((((-(...((((...))))((.(((..........((((((((((((.....)).))))))))))(((((.......)))))....)))...))))))).)......---- ( -29.90, z-score = -0.98, R) >droEre2.scaffold_4845 17627257 94 + 22589142 AGGGCCUC--AAGGGGCGUGGCCGCUGAUCACAAAGGAGAUCUUAAUUGCCGCUUAACGAGCGAUUGAGCGCUCUGUUCAAGGGAGUGGCGCAUAU-------------------- ...((((.--...))))(((((((((.........((((..(((((((((((.....)).)))))))))..)))).........)))))).)))..-------------------- ( -34.97, z-score = -1.35, R) >droAna3.scaffold_12916 11088406 90 + 16180835 -----GAUAAAGGAGGCAUGACCUAUGAUCACAAAGGAGAUCUUAAUUGUCGAUUAA-GAGCAAUUGAAGCCUC---GUAGAGGAGUAGAAAUCUGAAA----------------- -----.......(((((.........((((........))))(((((((((......-)).))))))).)))))---.((((..........))))...----------------- ( -19.10, z-score = -1.02, R) >consensus AGGGCCUCAAAAGGGGCGUGACCCGUGAUCACAAAGGAAAUCUUAAUUGCCGCUUAACGAGCAAUUGAGGACUCUAUUCAAGGGAGUGGCGGAUACAGCUAAAGACAUACUU____ ...(((((....)))))((((.......))))........((((((((((((.....)).))))))))))(((((.......)))))............................. (-19.54 = -20.35 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:55 2011