
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,314,504 – 19,314,583 |
| Length | 79 |
| Max. P | 0.592403 |

| Location | 19,314,504 – 19,314,583 |
|---|---|
| Length | 79 |
| Sequences | 9 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
| Shannon entropy | 0.39278 |
| G+C content | 0.57886 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19314504 79 + 23011544 -------CCUCGCCCCUGCGAAA-UGCAAACCAAUUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGCGU--ACAUGGACCC- -------..((((....))))..-......(((...(((((((.(((.(.((((....)))).).))).))))))).--...)))....- ( -25.90, z-score = -1.92, R) >droSim1.chr2L 19006407 79 + 22036055 -------CCUCGCCCCUGCGAAA-UGCAAACCAAAUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGCGU--ACAUGGACCC- -------..((((....))))..-......(((...(((((((.(((.(.((((....)))).).))).))))))).--...)))....- ( -25.90, z-score = -2.00, R) >droSec1.super_7 2942962 79 + 3727775 -------CCUGGCCCCUGCGAAA-UGCAAACCAAUUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGCGU--ACAUGGACCC- -------...((....(((....-.)))..(((...(((((((.(((.(.((((....)))).).))).))))))).--...))).)).- ( -23.90, z-score = -0.70, R) >droYak2.chr2R 5822076 79 + 21139217 -------CCCCGCCCCUGCGAAA-UGCAAACCAAUUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGCGU--ACAUGGGCUC- -------....((((.(((....-.))).........((((((.(((.(.((((....)))).).))).))))))..--....))))..- ( -27.90, z-score = -1.73, R) >droEre2.scaffold_4845 17619725 77 - 22589142 -------CCUCGCCCCUGCGAAA-UGCAAACCAAUUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGC----ACAUGGGCUC- -------....((((.(((....-.)))........(((((((.(((.(.((((....)))).).))).))))))----)...))))..- ( -28.50, z-score = -2.35, R) >droAna3.scaffold_12916 11081611 81 - 16180835 ------CCCCAUCCCCCGAGACA-UGCAAACCAAUUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGCGG--ACAUGGGCCGC ------.(((((...(((.....-.(((((....)))))((((.(((.(.((((....)))).).))).)))).)))--..))))).... ( -26.70, z-score = -0.89, R) >dp4.chr4_group4 399960 63 - 6586962 -------------------GAAA-UGCAAACCAAUUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGCGG--ACAUGG----- -------------------....-......(((.(((((((((.(((.(.((((....)))).).))).))))))))--)..)))----- ( -22.60, z-score = -2.15, R) >droWil1.scaffold_180703 152714 79 + 3946847 -------CCACCCCUUCGCGAAACUGCAAACCAAUUUGCGGCAAGUGCCUCGGUUGCCA---UGACACAUGCCGCGCGGACAUGGUCAC- -------..(((..((((((.....(((((....)))))((((.(((.(..((...)).---.).))).)))).))))))...)))...- ( -24.00, z-score = -0.64, R) >droVir3.scaffold_12723 3251077 79 + 5802038 CCUCGAAGCGAUCCUCAGUGAAA-UGCAAACCAAUUUGUGGCAAGUGCCUCGGUUGCUA---UGACACAUACGGUGC--CCGUGG----- ..(((.(((((((..........-.(((((....)))))(((....)))..))))))).---)))....(((((...--))))).----- ( -19.00, z-score = 0.40, R) >consensus _______CCUCGCCCCUGCGAAA_UGCAAACCAAUUUGCGGCAAGUGACUCGGUUGCCAUCGUGACACAUGCCGCGU__ACAUGGACCC_ ..............................(((...(((((((.(((.(.((((....)))).).))).)))))))......)))..... (-16.14 = -16.94 + 0.80)
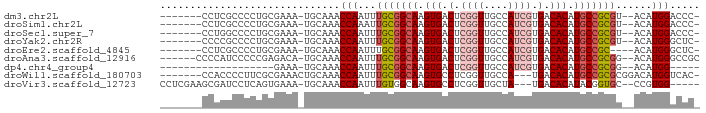

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:53 2011