| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,279,006 – 19,279,114 |
| Length | 108 |
| Max. P | 0.505853 |

| Location | 19,279,006 – 19,279,114 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Shannon entropy | 0.51207 |
| G+C content | 0.41623 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.59 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
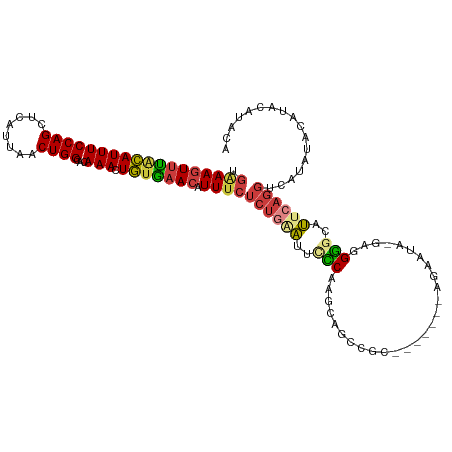
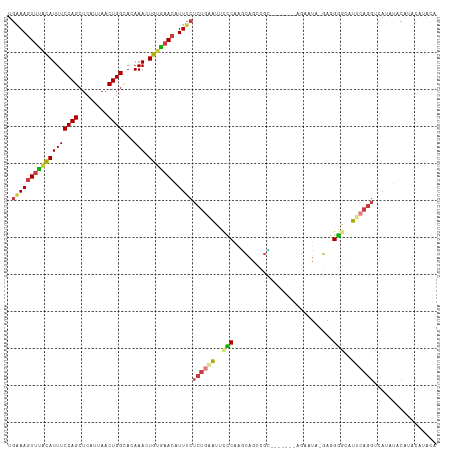
>dm3.chr2L 19279006 108 - 23011544 UGAAAGUUUACAUUUCCAGCUCAUUAACUGGCACAAACUGUGAACAUUUCUCUGAAUUCCCAAACAGUCGU-------AGAAUA-GAGGGACCUUCAGGUCAUAUACAUACAUAUA .((((((((((((((((((........))))...))).))))))).))))((((((.((((..........-------......-..))))..))))))................. ( -20.67, z-score = -0.80, R) >droSim1.chr2L 18970013 108 - 22036055 UGAAAGUUUACAUUUCCAGCUCAUUAACUGGCACAAACUGUGAACAUUUCUCUGAAUUCCCAAGCAGCCGU-------AGAAUA-GAGGGGCCUUCAGGUCAUAUACAUACAUACC .((((((((((((((((((........))))...))).))))))).))))((((((..(((...(......-------......-)..)))..))))))................. ( -20.70, z-score = -0.13, R) >droSec1.super_7 2907569 108 - 3727775 UGAAAGUUUACAUUUCCAGCUCAUUAACUGGCACAAACUGUGAACAUUUCUCUGGAUUCCCAAGCAGCCGU-------AGAAUA-GAGGGGCCUUCAGGUCAUAUACAUACAUACC .((((((((((((((((((........))))...))).))))))).))))..(((....))).......((-------(..(((-....((((....)))).)))...)))..... ( -20.40, z-score = 0.35, R) >droYak2.chr2R 5786842 108 - 21139217 UGAAAGUUCACAUUUCCAGCUCAUUAACUGGUACAAACUGUGAACAUUUCUCUGAAUUCCCAAGCAGCCGC-------AAAAUA-GAGGGGCAUUCAGGCCAUAUACAUACAUAUA .(((((((((((...((((........)))).......))))))).))))(((((((.(((..((....))-------......-...))).)))))))................. ( -24.60, z-score = -1.82, R) >droEre2.scaffold_4845 17585818 115 + 22589142 UGAAAGUUCGCAUUUCCAGCUCAUUAACUGGCACAAACUGUGAACAUUUCUCUGAAUUUCCAAGCAGCCGCAGAACAGAGAAUA-GAAGGGCAUUCAGGCCACGCACAUACACAUA .....((((((((((((((........))))...))).)))))))..(((((((...(((...((....)).))))))))))..-(...(((......)))...)........... ( -25.10, z-score = -0.86, R) >droAna3.scaffold_12916 11040223 107 + 16180835 UGGAAGUUUACAUUUCCAGCUCAUUAACUGGCACAAACUGUGAACAUUUCUCU-AAUUCUCAUCCAGCCGG-------GGACUACUAAGAGUAUAAAGGGCACAUUCCGCCCAAC- (((((((....)))))))((((.....(((((.......((((..(((.....-)))..))))...)))))-------((....))..)))).....((((.......))))...- ( -23.30, z-score = -0.91, R) >droVir3.scaffold_12723 3217999 81 - 5802038 AAAAACUUUAUAUUUCCAGCUCAUUAACUGGGACAAACUACACACAUUGC-----GUACUCGGGAAGUCCC--------CAACUCUAAGGCUAC---------------------- .....(((((...((((((........)))))).................-----......(((.....))--------).....)))))....---------------------- ( -11.90, z-score = -0.30, R) >consensus UGAAAGUUUACAUUUCCAGCUCAUUAACUGGCACAAACUGUGAACAUUUCUCUGAAUUCCCAAGCAGCCGC_______AGAAUA_GAGGGGCAUUCAGGUCAUAUACAUACAUACA .((((((((((((((((((........))))...))).))))))).))))((((((..(((...........................)))..))))))................. (-13.63 = -13.59 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:48 2011