
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,276,594 – 19,276,714 |
| Length | 120 |
| Max. P | 0.812664 |

| Location | 19,276,594 – 19,276,692 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.07 |
| Shannon entropy | 0.50040 |
| G+C content | 0.45968 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -12.07 |
| Energy contribution | -12.17 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
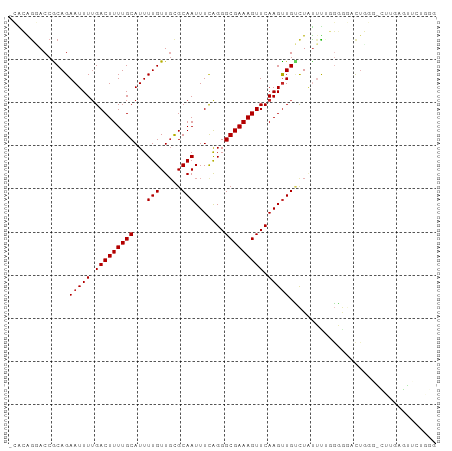
>dm3.chr2L 19276594 98 + 23011544 -CACAGGACCACAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUUGGGGGACUGGGACUCGAGUUCUGGG -..(((..((.((((((..((((((..(((......))).........((((....))))))))))...)))))).))..)))((((....)))).... ( -27.90, z-score = -1.57, R) >droSim1.chr2L 18967608 98 + 22036055 -CACAGGACCACAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUCGGGGGACUGGGACUUGAGUUCUGGG -..((((((((.(((..((((((((((((...(((.....))).......))))))).)))))...))).(..(((....)))..)..)).)))))).. ( -29.00, z-score = -1.95, R) >droSec1.super_7 2905213 98 + 3727775 -CACAAUACCACAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUCGGGGGACUGGGACUUGAGUUCUGGG -.......((.((((((((........(((......)))(((((((..((((....)))))))))))...(..(((....)))..)...)))))))))) ( -28.00, z-score = -2.15, R) >droYak2.chr2R 5784340 98 + 21139217 -CACAGGACCACAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUUCGGGGGCCUGGGCUUGAGUUCUGGG -..((((.((.(((..((((((.....(((......)))(((((((..((((....)))))))))))......))))))..)))))))))......... ( -25.30, z-score = -0.13, R) >droEre2.scaffold_4845 17583544 97 - 22589142 -CACAGGACCGCAGAAUUUUGACUUUUGCAUUUUGUUGUGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUCGGGGG-CUUGGGCUUGAGUUCUGGG -..(((((((((.....(((((...((((((......))))))..)))))))).(((((((((((.((......)).))-)))))))))..)))))).. ( -32.00, z-score = -2.65, R) >droAna3.scaffold_12916 11037858 98 - 16180835 UCAAAGGACCGCAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUGUUCUGGUCU-UUUCAGUACCAGUUCUGGG ..(((((((((((((..((((((((((((...(((.....))).......))))))).)))))...)))))...)))))-))).....(((....))). ( -29.20, z-score = -2.17, R) >dp4.chr4_group4 355734 83 - 6586962 -GGCGGCACCGCAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCGGGGCGAAAGUUCAAGUUGCCUGUUCUGGGGUUUUGU--------------- -.((((.(((.((((((..........(((......)))((((((...((((....)))).))))))..)))))).))).))))--------------- ( -28.10, z-score = -1.82, R) >droPer1.super_16 665137 83 + 1990440 -GGCGGCACCGCAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCGGGGCGAAAGUUCAAGUUGCCUGUUCUGGGGUUUUGU--------------- -.((((.(((.((((((..........(((......)))((((((...((((....)))).))))))..)))))).))).))))--------------- ( -28.10, z-score = -1.82, R) >droVir3.scaffold_12723 3215825 85 + 5802038 CAGAGGCACCGCAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUUGGGGCGAAAGUUCAAGCUGGUUUCGCAUUCAGCUUGGC-------------- (((((....(((((.....((.(....))).....)))))...)))))((((....))))(((((((........)))))))...-------------- ( -22.00, z-score = 0.19, R) >droGri2.scaffold_15126 8138100 97 - 8399593 UAGAGGCACCGCAGAAUUU-GACUUUUGCAUUUUGUUUCGCAAUUUCGGGGCGAAAGUUCAAGUUGGUUCCGUAUUCAGCUUUGCUUCUCCAGUCUGU- .(((((((..((.(((((.-.((((..((.((((((..((......))..))))))))..))))..)......)))).))..))))))).........- ( -27.00, z-score = -1.45, R) >consensus _CACAGGACCGCAGAAUUUUGACUUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUUGGGGGACUGGG_CUUGAGUUCUGGG ..............(((((.(((((((((...(((.....))).......))))))))).))))).................................. (-12.07 = -12.17 + 0.10)

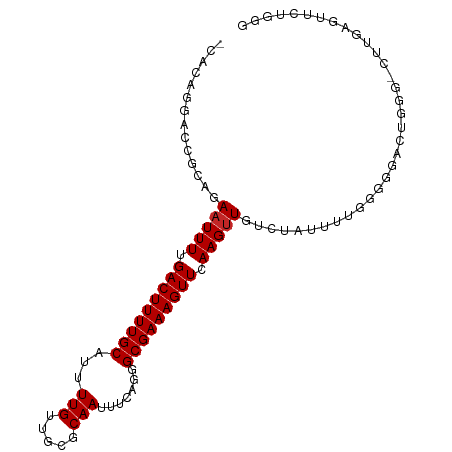
| Location | 19,276,616 – 19,276,714 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 98.64 |
| Shannon entropy | 0.01874 |
| G+C content | 0.47959 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19276616 98 + 23011544 UUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUUGGGGGACUGGGACUCGAGUUCUGGGUUUGCUGCCGGCUUGUUUCUGC ....(((......)))((((.((((((((...(((((.((((.(((.....))).)))).)))))...)))))))).))))................. ( -28.70, z-score = -0.97, R) >droSim1.chr2L 18967630 98 + 22036055 UUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUCGGGGGACUGGGACUUGAGUUCUGGGUUUGCUGCCGGCUUGUUUCUGC ....(((......)))((((.((((((((..((((((.((((.(((.....))).)))).))))))..)))))))).))))................. ( -31.00, z-score = -1.76, R) >droSec1.super_7 2905235 98 + 3727775 UUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUCGGGGGACUGGGACUUGAGUUCUGGGUUUGCUGCCGGCUUGUUUCUGC ....(((......)))((((.((((((((..((((((.((((.(((.....))).)))).))))))..)))))))).))))................. ( -31.00, z-score = -1.76, R) >consensus UUUUGCAUUUUGUUGCGCAAUUUCAGGGCGAAAGUUCAAGUUGUCUAUUUCGGGGGACUGGGACUUGAGUUCUGGGUUUGCUGCCGGCUUGUUUCUGC ....(((......)))((((.((((((((..((((((.((((.(((.....))).)))).))))))..)))))))).))))................. (-29.30 = -29.63 + 0.33)


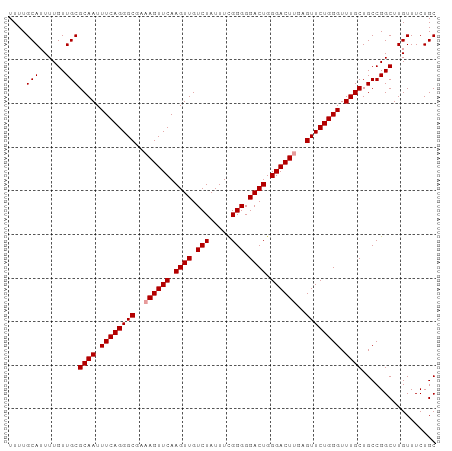
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:46 2011