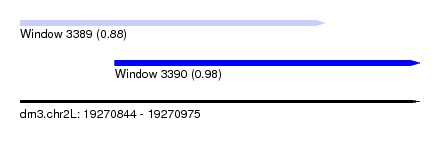
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,270,844 – 19,270,975 |
| Length | 131 |
| Max. P | 0.978982 |

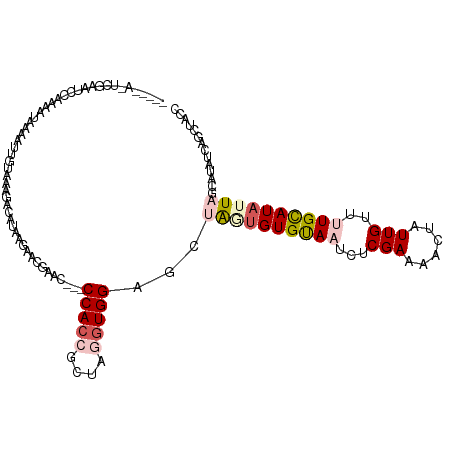
| Location | 19,270,844 – 19,270,944 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 58.15 |
| Shannon entropy | 0.74510 |
| G+C content | 0.37169 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -8.58 |
| Energy contribution | -9.94 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19270844 100 + 23011544 -----------AUUCAA-AUCGGAUUCUUCAUUCAU---AAGGUAC--CCCACCGCUAGGUGGAGCUGAUGUGUAAUCUCGAUAACUAUUGUUUUGUAUAUUAGCAUAUCAGCUACC -----------......-...((((.....))))..---..(((..--.(((((....)))))((((((((((((((..(((.(((....))))))...))).)))))))))))))) ( -27.80, z-score = -3.06, R) >droSim1.chr2L 18962013 112 + 22036055 -----AUUCGAAUCGAAAUAUUCAUUAAAAAGUUGUAAAAACAAGUUACCCACAGCUAGGUGGCGCUAGUGUGUAAUCUCGAAAAUCAUUGUUUUGCAUUUUAGCAUAUCAGCUACC -----....((((......)))).......(((((.........((((((........))))))(((((.((((((...(((......)))..)))))).)))))....)))))... ( -23.30, z-score = -1.80, R) >droSec1.super_7 2899725 108 + 3727775 -------UUGUUUUCAAUAUCAAAUUUCAAAUACAUGAUAACAAUC--ACCACCGCUAGGUGGCGCUAGUGUGUAAUCUCGAAAACUAUUGUUUUGUAUAUUAUCAUAUCAGCUACC -------(((((.(((...................))).)))))..--.(((((....))))).(((.(((((((((....(((((....)))))....)))).))))).))).... ( -21.21, z-score = -2.01, R) >droYak2.chr2R 5778553 105 + 21139217 ----AAACCGAUGCAUUAAUAAAAUUGUAAUGACAAACGAAUGUAC---CCAAAGCUAGGUGGGGCUACUGUGUAAUAUCGAACACUAUUGCUGUGCAUAUUUUCAGCUACC----- ----......((((((........((((....)))).........(---(((........))))((....((((........))))....)).)))))).............----- ( -16.60, z-score = 1.14, R) >droEre2.scaffold_4845 17578010 101 - 22589142 GAACAAAUGAACAAGUUCAUAAAAGAGUUUUGACAAUUGAAUGCAC---CCACCGCUAGGUGGAGCUAUUGUGCAAUCUCGAAAACUGUUGCGCUGCAUAUUUC------------- ......(((((....)))))....................(((((.---(((((....))))).......(((((((..........)))))))))))).....------------- ( -22.90, z-score = -0.85, R) >consensus _____A_UCGAAUCCAAAAUAAAAUUGUAAAGACAUAAGAACGAAC___CCACCGCUAGGUGGAGCUAGUGUGUAAUCUCGAAAACUAUUGUUUUGCAUAUUAGCAUAUCAGCUACC .................................................(((((....)))))...((((((((((...(((......)))..)))))))))).............. ( -8.58 = -9.94 + 1.36)

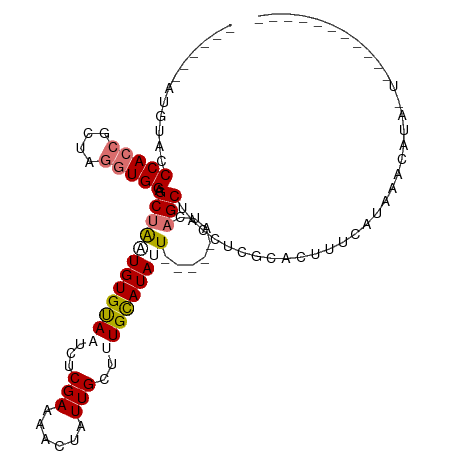
| Location | 19,270,875 – 19,270,975 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 60.66 |
| Shannon entropy | 0.63189 |
| G+C content | 0.40618 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -9.59 |
| Energy contribution | -10.65 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19270875 100 + 23011544 -----------CCCACCGCUAGGUGGAGCUGAUGUGUAAUCUCGAUAACUAUUGUUUUGUAUAUUAGCAUAUCAGCUACCUCGCACUUUCAUAAUCAAAGU-CGAAUAACUA -----------.(((((....)))))((((((((((((((..(((.(((....))))))...))).)))))))))))...(((.(((((.......)))))-)))....... ( -30.00, z-score = -4.29, R) >droSec1.super_7 2899753 112 + 3727775 UGAUAACAAUCACCACCGCUAGGUGGCGCUAGUGUGUAAUCUCGAAAACUAUUGUUUUGUAUAUUAUCAUAUCAGCUACCUCGCACAUUCAUAACUAUAAUAUUAAUUUCUU ((((....)))).....((.(((((((....(((((((((....(((((....)))))....)))).)))))..))))))).))............................ ( -21.50, z-score = -1.97, R) >droYak2.chr2R 5778589 85 + 21139217 ------AUGUACCCAAAGCUAGGUGGGGCUACUGUGUAAUAUCGAACACUAUUGCUGUGCAUAUU-----UUCAGCUACCUAGCACUUUCAUAAGC---------------- ------...........((((((((((((....((((........))))....))).((......-----..)).)))))))))............---------------- ( -20.40, z-score = -0.28, R) >droEre2.scaffold_4845 17578050 89 - 22589142 ------AUGCACCCACCGCUAGGUGGAGCUAUUGUGCAAUCUCGAAAACUGUUGCGCUGCAUAUU-------UCGCAGUUUCACAAGCUGAUCGAAAUACUG---------- ------.((((.(((((....))))).......(((((((..........)))))))))))((((-------((((((((.....)))))..)))))))...---------- ( -27.30, z-score = -2.26, R) >consensus ______AUGUACCCACCGCUAGGUGGAGCUAAUGUGUAAUCUCGAAAACUAUUGCUUUGCAUAUU_____AUCAGCUACCUCGCACUUUCAUAAACAUA_U___________ ............(((((....))))).((((((((((((...(((......)))..)))))))))........))).................................... ( -9.59 = -10.65 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:42 2011