| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,265,610 – 19,265,709 |
| Length | 99 |
| Max. P | 0.637887 |

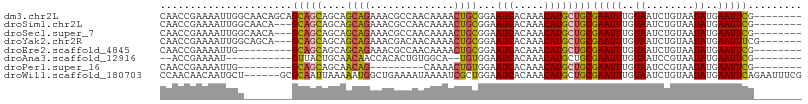
| Location | 19,265,610 – 19,265,709 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Shannon entropy | 0.43051 |
| G+C content | 0.41151 |
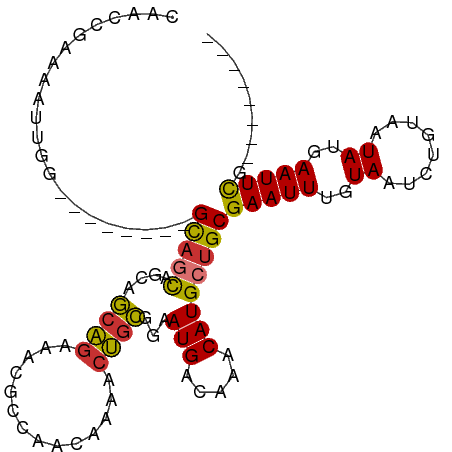
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.03 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19265610 99 - 23011544 CAACCGAAAAUUGGCAACAGCAGCAGCAGCAGCAGAAACGCCAACAAAACUGCGGAAUGACAAACAUGCUGCGAAUUUGUAAUCUGUAAUAUGAAUUCG-------- ...........((....))((((((.......((....(((..........)))...)).......))))))(((((..((........))..))))).-------- ( -22.34, z-score = -0.91, R) >droSim1.chr2L 18955718 96 - 22036055 CAACCGAAAAUUGGCAACA---GCAGCAGCAGCAGAAACGCCAACAAAACUGCGGAAUGACAAACAUGCUGCGAAUUUGUAAUCUGUAAUAUGAAUUCG-------- ..........(((....))---)..((((((((((..............))))(......).....))))))(((((..((........))..))))).-------- ( -21.84, z-score = -1.28, R) >droSec1.super_7 2894455 96 - 3727775 CAACCGAAAAUUGGCAACA---GCAGCAGCAGCAGAAACGCCAACAAAACUGCGGAAUGACAAACAUGCUGCGAAUUUGUAAUCUGUAAUAUGAAUUCG-------- ..........(((....))---)..((((((((((..............))))(......).....))))))(((((..((........))..))))).-------- ( -21.84, z-score = -1.28, R) >droYak2.chr2R 5773226 97 - 21139217 CAACCGAAAAUUGGCAGCA---GCAGCAGCAGCAGAAACGACAACAAAACUGCGGAAUGACAAACAUGCUGCGAAUUUGUAAUCUGUAAUAUGAAUUUCG------- ....((((((((.((((((---((....)).((((..............)))).............)))))).)))))....((........))...)))------- ( -19.74, z-score = -0.28, R) >droEre2.scaffold_4845 17572765 90 + 22589142 CAACCGAAAAUUG---------GCAGCAGCAGCAGAAACGCCAACAAAACUGCGGAAUGACAAACAUGCUGCGAAUUUGUAAUCUGUAAUAUGAAUUCG-------- ...((((...)))---------)..((((((((((..............))))(......).....))))))(((((..((........))..))))).-------- ( -20.24, z-score = -1.37, R) >droAna3.scaffold_12916 11026254 84 + 16180835 --ACCGAAAAU-----------GUUACUGCAACAACCACACUGUGGCA--UGUGGAAUGACAAACAUGCUGCGAAUUUGUAAUCCGUAAUAUGAAUUCG-------- --..((((.((-----------(((((.(..((((......((..(((--(((..........))))))..))...))))...).)))))))...))))-------- ( -19.00, z-score = -1.39, R) >droPer1.super_16 654498 81 - 1990440 CAACCGAAAAUUG---------GCAGCAGCAACAG---------CAAAACUGUGGAAUGACAAACAUGCUGCGAAUUUGUAAUCCGUAAUAUGAAUUCG-------- ...((((...)))---------)..((((((((((---------.....))))(......).....))))))(((((..((........))..))))).-------- ( -18.60, z-score = -1.63, R) >droWil1.scaffold_180703 3724631 101 + 3946847 CCAACAACAAUGCU------GCGCAAUUAAAAAUGGCUGAAAAUAAAAUCGCUGGAAUGACAAACAUGCUGCGAAUUUGUAAUCUGUAAUAUGAAUUCAGAAUUUCG ..............------((.((........)))).(((((((((.((((.((.(((.....))).)))))).)))))..((((.(((....))))))).)))). ( -14.00, z-score = 0.63, R) >consensus CAACCGAAAAUUGG________GCAGCAGCAGCAGAAACGCCAACAAAACUGCGGAAUGACAAACAUGCUGCGAAUUUGUAAUCUGUAAUAUGAAUUCG________ ......................(((((....((((..............))))...(((.....))))))))(((((..((........))..)))))......... (-14.42 = -14.03 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:41 2011