| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,248,984 – 19,249,076 |
| Length | 92 |
| Max. P | 0.702407 |

| Location | 19,248,984 – 19,249,076 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 59.96 |
| Shannon entropy | 0.80646 |
| G+C content | 0.50510 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -7.89 |
| Energy contribution | -7.82 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.702407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19248984 92 + 23011544 -----CGCGCC--UUGCCGCAUAAAAUGUGGCACAUUCCCAGACGAUGGGGAUACUAA-GGGAUACAGAUCUAG--AUACGCGUACAAAUACACACACGCCG-- -----((((((--(((((((((...)))))))..(((((((.....)))))))...))-))(((....)))...--...))))...................-- ( -26.30, z-score = -2.34, R) >droSim1.chr2L 18939135 92 + 22036055 -----CGCGCC--UUGCCGCAUAAAAUGUGGCACAUUCCCAGACGAUGGGGAUACUAA-GGGAUACAGAUACAG--AUACGCGCACAAAUACACACACAGCG-- -----.((((.--.((((((((...)))))))).(((((((.....))))))).....-...............--....))))..................-- ( -26.20, z-score = -3.10, R) >droSec1.super_7 2872353 92 + 3727775 -----CGCGCC--UUGCCGCAUAAAAUGUGGCACAUUCCCAGACGAUGGGGAUACUAA-GGGAUACGGAUACAG--AUACGCGUACAAAUACACACACAACG-- -----((((((--(((((((((...)))))))..(((((((.....)))))))...))-)).....(....)..--...))))...................-- ( -25.40, z-score = -2.73, R) >droYak2.chr2R 5756629 88 + 21139217 -----CGCGCC--UUGCCGCAUAAAAUGUGGCACAUUCCCAGACGAUGGGGAUACCAA-GGGAUACUAA--GGG--AUACAGAUACAAAUGCACACAGAG---- -----.(((((--(((((((((...)))))))..(((((((.....)))))))...))-))........--(..--...).........)))........---- ( -22.80, z-score = -1.80, R) >droEre2.scaffold_4845 17556579 92 - 22589142 -----CGCGCC--UUGCCGCAUAAAAUGUGGCACAUUCCCAGACGAUGGGGGCACUAA-GGGAUACAGAUACAG--AUGCAGAUGCAGAUACACACGGAGGA-- -----...(((--.((((((((...))))))))....((((.....))))))).....-...............--.(((....)))...............-- ( -23.10, z-score = -0.56, R) >droAna3.scaffold_12916 11002049 95 - 16180835 -----CUCGCC--UUGCCGCAUAAAAUGUGGCACAUUCCCAGACGGGCAACACGGAAAAGGGACAGGGCUACAGGGAUAGUGAAAUAGAUAGAGAUGGAGGG-- -----((((((--(((((((((...)))))))....((((...(((....).)).....)))).)))))(((.......))).........)))........-- ( -26.40, z-score = -1.63, R) >dp4.chr4_group4 330649 103 - 6586962 CUCGCCUAGCCAGCGGCCGCAUAAAAUGUGGCACAUUCCCAGACGGCCAGCGAAAAGA-GAUAUGUAUGUAUAUAUGUAUAUGUGUGAACAAUAGCGCGGGGGG ((((((((....((((((((....((((.....))))....).))))).)).......-..((..(((((((....)))))))..)).....))).)))))... ( -28.70, z-score = 0.21, R) >droPer1.super_16 640519 103 + 1990440 CUCGCCUAGCCAGCGGCCGCAUAAAAUGUGGCACAUUCCCAGACGGCCAGCGAAAAGA-GAUAUGUAUGUAUAUAUGUAUAUGUGAGAACAAUAGCGCGGGGGG ((((((((....((((((((....((((.....))))....).))))).)).......-.((((((((((....))))))))))........))).)))))... ( -28.10, z-score = 0.07, R) >droWil1.scaffold_180703 3707022 90 - 3946847 -----CAAUUCUCCAUAUAUAAAUUGUGUGGCAUAUACCCAGCCAGCACCAUCAUCAUGACCAGAACAUCCCAG--AGGCCAGAUUAUCCGGGAGUA------- -----.....((((...........(((((((.........)))).)))....(((.((.((............--.)).)))))......))))..------- ( -14.42, z-score = 0.86, R) >consensus _____CGCGCC__UUGCCGCAUAAAAUGUGGCACAUUCCCAGACGAUGGGGAUACUAA_GGGAUACAGAUACAG__AUACACGUACAAAUACACACACAGGG__ ...............((((((.....))))))........................................................................ ( -7.89 = -7.82 + -0.07)


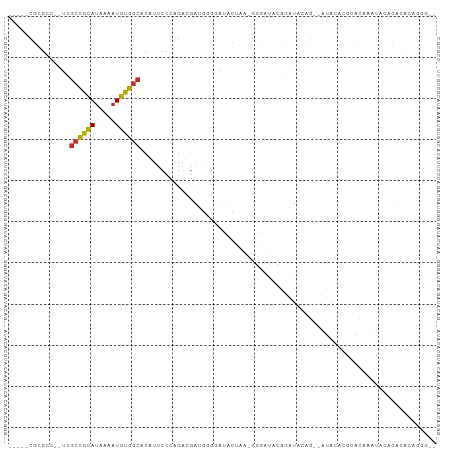
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:38 2011