| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,233,799 – 19,233,895 |
| Length | 96 |
| Max. P | 0.990338 |

| Location | 19,233,799 – 19,233,895 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.51693 |
| G+C content | 0.47535 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -16.63 |
| Energy contribution | -16.31 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19233799 96 - 23011544 ------------GAAAGUGGCUGGGGC--AAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCUCCGCCGCCCUUCCUUUCCUAUUUUUU ------------(((.(((((.(((((--(((((((((((((((...........)))))...)))))))))........)))))))))))..))).............. ( -30.60, z-score = -2.87, R) >droPer1.super_16 622743 95 - 1990440 ------------GGAAGGGGAAAGGGC--AAAGCGCAU-AAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCCCGGCCCCUCUUCAGUUCCCCGUUCCC ------------((((((((((.((((--(((((((((-(((((...........))))....)))))))))........)))))(....).......)))))).)))). ( -33.70, z-score = -2.40, R) >dp4.chr4_group4 313009 95 + 6586962 -------------GGAAGGGGCAGGGC--AAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCCCGGCCCCUCUUCAGUUCCCCGUUCCC -------------(((((((((.((((--(((((((((((((((...........)))))...)))))))))........))))).)))))).))).............. ( -36.10, z-score = -2.88, R) >droAna3.scaffold_12916 10985019 96 + 16180835 ------------AAAAGGGGGCGGGGCUGAAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACCGCCU--CUUUCCUCCACUCCCUAUUUUUU ------------..((((((((((.((...((((((((((((((...........)))))...)))))))))...))..))))))--))))................... ( -31.50, z-score = -2.80, R) >droEre2.scaffold_4845 17541316 96 + 22589142 ------------GAAAGUGGCUGGGGC--AAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCUCCGCCGCCCUUCCUGCCCCGCUUUUU ------------(((((((((.(((((--.((((((((((((((...........)))))...)))))))))...(((.......))).)))))....)))..)))))). ( -31.50, z-score = -2.18, R) >droYak2.chr2R 5741519 95 - 21139217 ------------GAAAGUGGCUGGGGC--AAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCUCCGCCGCCCUUCUU-CCCUAUUUUUU ------------(((.(((((.(((((--(((((((((((((((...........)))))...)))))))))........)))))))))))..)))..-........... ( -31.00, z-score = -3.04, R) >droSec1.super_7 2857099 96 - 3727775 ------------GAAAGUGGCUGGGGC--AAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCUCCGCCGCCCUUCCUUCCCUAUUUUUU ------------(((.(((((.(((((--(((((((((((((((...........)))))...)))))))))........)))))))))))..))).............. ( -30.60, z-score = -2.81, R) >droSim1.chr2L 18924254 96 - 22036055 ------------GAAAGUGGCUGGGGC--AAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCUCCGCCGCCCUUCCUUCCCUAUUUUUU ------------(((.(((((.(((((--(((((((((((((((...........)))))...)))))))))........)))))))))))..))).............. ( -30.60, z-score = -2.81, R) >droGri2.scaffold_15126 8098714 76 + 8399593 -------------------GUCGGGGGCCAAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCCCCUCUCUCUCA--------------- -------------------(..((((((((((((((((((((((...........)))))...)))))))))....))...))))))..).....--------------- ( -22.90, z-score = -2.38, R) >droWil1.scaffold_180703 3687142 107 + 3946847 GGGAAGGGCAUGGAAGGCAGAACGGGGCCAAAGAGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCCUCCUGCUGCAG-GGCUCGCUCUUU-- ..(((((((..(((.(((((.((((.....(((.((((((((((...........)))))...))))).))).))))..)))))))).(((....-)))..)))))))-- ( -31.50, z-score = -0.20, R) >droMoj3.scaffold_6500 25683600 91 + 32352404 -------------------GUCGGGGGCUAAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGUAUGCCACGCCCUCCCGCUGCCCGCCCCGCGC -------------------((.((((((..((((((((((((((...........)))))...)))))))))...(((.....))))))).)).))....(((...))). ( -28.10, z-score = -0.57, R) >consensus ____________GAAAGUGGCUGGGGC__AAAGCGCAUAAAAAGGAUAUUUGACACUUUUAAAUGUGCGCUUAUUGUGACUGCCCCGCCCCCCUUCCUUCCCCAUUUUUU ......................(((((...((((((((((((((...........)))))...)))))))))...((....))......)))))................ (-16.63 = -16.31 + -0.32)

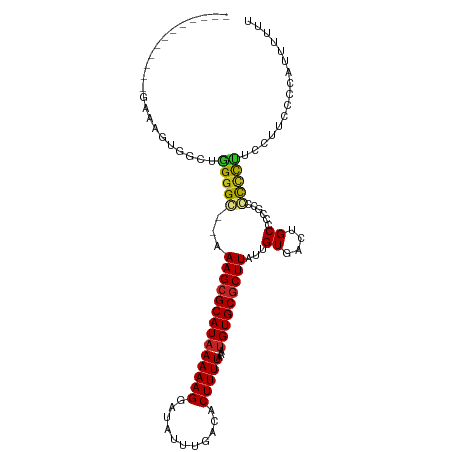
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:35 2011