| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,223,051 – 19,223,150 |
| Length | 99 |
| Max. P | 0.505913 |

| Location | 19,223,051 – 19,223,150 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Shannon entropy | 0.49364 |
| G+C content | 0.47058 |
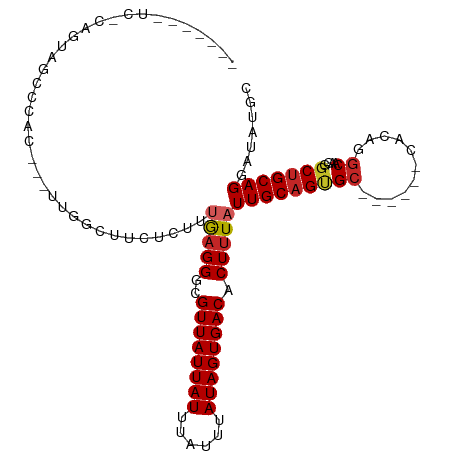
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19223051 99 + 23011544 ------GUUCGCACAGUAUCCCAGUUGGCUUCUCUUUGAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACAGGCAACGGCUGCAGGAUAUGC ------.........((((((((((((((..(((..(((((..((((((((......)))))))).)))))..).)).))------)...(....))))))..)))))).. ( -28.00, z-score = -1.26, R) >dp4.chr4_group4 302217 89 - 6586962 -------------UAGCUCUC---UCAGCAUCAGUUUAAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACCGGCGGCGGCUGCAGGAUACGC -------------..(((((.---..(((....)))..)))))((((((((......))))))))......(((((((((------(......))).)))))))....... ( -26.40, z-score = -1.55, R) >droPer1.super_16 611923 89 + 1990440 -------------UAGCUCUC---UCAGCAUCAGUUUAAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACCGGCGGCGGCUGCAGGAUACGC -------------..(((((.---..(((....)))..)))))((((((((......))))))))......(((((((((------(......))).)))))))....... ( -26.40, z-score = -1.55, R) >droWil1.scaffold_180703 3662275 87 - 3946847 ------GUGUGCACAGCAUGC---UUGGCCU---UCGAAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUG----------AAGC--CGCCGGCAGGAUACGC ------((((((((.(((...---...((((---.....))))((((((((......)))))))).......))).)))----------..((--(...)))....))))) ( -23.20, z-score = 0.01, R) >droSim1.chr2L 18913547 102 + 22036055 GUUCGUAUCCCAGUAUCCCAG---UUGGCUUCUCUUUGAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACAGGCAACGGCUGCAGGAUAUGC ...(((((((((((...((.(---(.(((..(((..(((((..((((((((......)))))))).)))))..).)).))------))).)).....))))..))))))). ( -29.90, z-score = -1.78, R) >droSec1.super_7 2846383 102 + 3727775 GUUCGUAUCCCAGUAUCCCAG---UUGGCUUCUCUUUGAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACAAGCAACGGCUGCAGGAUAUGC ...(((((((((((.......---.((((..(((..(((((..((((((((......)))))))).)))))..).)).))------)).........))))..))))))). ( -28.33, z-score = -1.58, R) >droYak2.chr2R 5730599 99 + 21139217 GUUC---UCACAGUAUCCCCG---UUGGCUUCUCUUUGAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACAGGCAACGGCUGCAGGAUAUGC ....---.....(((((((((---(((.((.(((...)))(((((((((((......))))))))(((.......)))))------)..)).)))))).....)))))).. ( -27.40, z-score = -1.28, R) >droEre2.scaffold_4845 17530783 99 - 22589142 GUUC---UCACAGUAUCCCCG---UUGGCGUCUCUUUGAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACAGGCAACGGCUGCAGGAUAUGC ((((---(..((((....((.---.(((((.(((..(((((..((((((((......)))))))).)))))..).)))))------))..)).....)))).))))).... ( -27.50, z-score = -1.11, R) >droAna3.scaffold_12916 10975194 100 - 16180835 --GCUCUUGUACGCUCCUUGC---UGGCUUCCCUUUUGAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC------CACCGGCAACGGCUGCAGGAUAUGC --..(((((((.((.(.((((---(((...((((....)))).((((((((......))))))))...............------..))))))).))))))))))..... ( -30.90, z-score = -1.69, R) >droMoj3.scaffold_6500 25671727 90 - 32352404 ------------------CAC---UCGGUGCGUAUUCGUGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGCGCAGCAGCCGCCGGCGCCGGCUGCAGGAUAUGC ------------------...---.....((((((((..(((.((((((((......)))))))).)))...((((((.(.((.(((...))))).))))))))))))))) ( -31.50, z-score = -0.87, R) >consensus _______UC_CAGUAGCCCAC___UUGGCUUCUCUUUGAGGGCGUUAUUAUUUAUUUAUAGUGACACUUUAUUGCAGUGC______CACAGGCAACGGCUGCAGGAUAUGC ....................................(((((..((((((((......)))))))).)))))(((((((...................)))))))....... (-14.21 = -14.42 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:33 2011