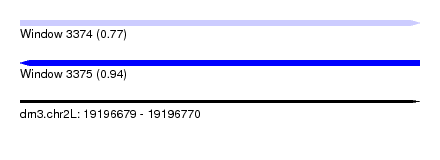
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,196,679 – 19,196,770 |
| Length | 91 |
| Max. P | 0.939982 |

| Location | 19,196,679 – 19,196,770 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 62.27 |
| Shannon entropy | 0.70310 |
| G+C content | 0.60049 |
| Mean single sequence MFE | -33.81 |
| Consensus MFE | -15.81 |
| Energy contribution | -15.71 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
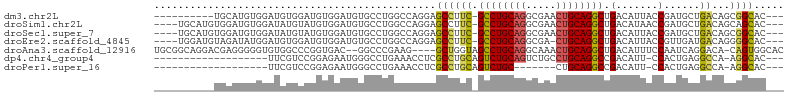
>dm3.chr2L 19196679 91 + 23011544 ----------UGCAUGUGGAUGUGGAUGUGGAUGUGCCUGGCCAGGAGCCUUC-GCCUGCAGGCGAACUGCAGGCUGACAUUACCGAUGCUGACAGCGGCAC--- ----------(((.(((((...((((((((((.((.(((....))).)).)))-((((((((.....))))))))..))))..)))...)).)))...))).--- ( -32.80, z-score = -0.11, R) >droSim1.chr2L 18886637 97 + 22036055 ----UGCAUGUGGAUGUGGAUAUGUAUGUGGAUGUGCCUGGCCAGGAGCCUUC-GCCUGCAGGCGAACUGCAGGCUGACAUAACCGAUGCUGACAGCAGCAC--- ----(((.(((((...(((.(((((....(((.((.(((....))).)).)))-((((((((.....))))))))..))))).)))...)).))))))....--- ( -36.10, z-score = -0.91, R) >droSec1.super_7 2820000 97 + 3727775 ----UGCAUGUGGAUGUGGAUAUGUAUGUGGAUGUGCCUGGCCAGGAGCCUUC-GCCUGCAGGCGAACUGCAGGCUGACAUUACCGAUGCUGACAGCGGCAC--- ----(((.(((((...(((..((((....(((.((.(((....))).)).)))-((((((((.....))))))))..))))..)))...)).)))...))).--- ( -33.80, z-score = -0.04, R) >droEre2.scaffold_4845 17505074 96 - 22589142 ----UGGAUGUAGAUAUGGAUGUGGAUGUGGAUGUGCCUGGCCAGGAGCCUUC-GCCUGCAGGCGA-CUGCAGGCUGACAUUACCGUUGAUGACAGGGGCAC--- ----....(((.(.(((.((((..((((((((.((.(((....))).)).)))-((((((((....-))))))))..)))))..)))).))).)....))).--- ( -33.40, z-score = -0.79, R) >droAna3.scaffold_12916 10949226 98 - 16180835 UGCGGCAGGACGAGGGGGUGUGGCCCGGUGAC--GGCCCGAAG----GCUGGUAGCCUGCAGGCAAACUGCAGGCUGACAUUUCCAAUCAGGACA-CAGUGGCAC (((.((.......((..((((((.(((....)--)).))....----.....((((((((((.....))))))))))))))..)).....(....-).)).))). ( -44.40, z-score = -2.03, R) >dp4.chr4_group4 274959 81 - 6586962 -------------------UUCGUCCGGAGAAUGGGCCUGAAACCUCGCCUGCAGUCUGCAGUCUGCCUGCAGGCCGACAUU-CCACUGAGGCCA-AGGCAC--- -------------------.....(((.....)))((((....((((((((((((...((.....)))))))))).(.....-.)...))))...-))))..--- ( -28.60, z-score = -0.44, R) >droPer1.super_16 584497 74 + 1990440 -------------------UUCGUCCGGAGAAUGGGCCUGAAACCUCGCCUGCAGUCUGC-------CUGCAGGCCGACAUU-CCACUGAGGCCA-AGGCAC--- -------------------.....(((.....)))((((....((((((((((((.....-------)))))))).(.....-.)...))))...-))))..--- ( -27.60, z-score = -1.18, R) >consensus __________UGGAUGUGGAUGUGCAUGUGGAUGUGCCUGGCCAGGAGCCUUC_GCCUGCAGGCGAACUGCAGGCUGACAUUACCGAUGAUGACAGCGGCAC___ ...............................................(((....((((((((.....)))))))).(.......)............)))..... (-15.81 = -15.71 + -0.10)
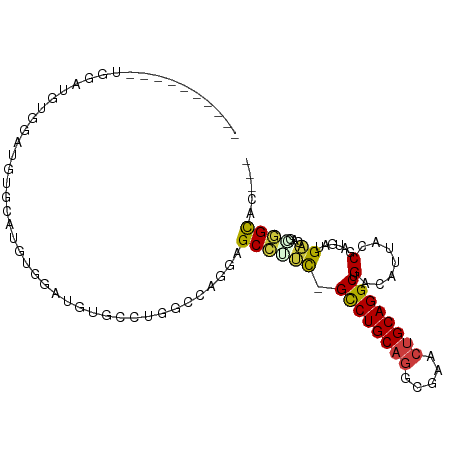
| Location | 19,196,679 – 19,196,770 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 62.27 |
| Shannon entropy | 0.70310 |
| G+C content | 0.60049 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.939982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19196679 91 - 23011544 ---GUGCCGCUGUCAGCAUCGGUAAUGUCAGCCUGCAGUUCGCCUGCAGGC-GAAGGCUCCUGGCCAGGCACAUCCACAUCCACAUCCACAUGCA---------- ---(((((((.....))...(((...(((.((((((((.....))))))))-...))).....))).))))).......................---------- ( -33.40, z-score = -1.32, R) >droSim1.chr2L 18886637 97 - 22036055 ---GUGCUGCUGUCAGCAUCGGUUAUGUCAGCCUGCAGUUCGCCUGCAGGC-GAAGGCUCCUGGCCAGGCACAUCCACAUACAUAUCCACAUCCACAUGCA---- ---((((((....)))))).((...((((.((((((((.....))))))))-...(((.....))).))))...)).........................---- ( -32.90, z-score = -0.90, R) >droSec1.super_7 2820000 97 - 3727775 ---GUGCCGCUGUCAGCAUCGGUAAUGUCAGCCUGCAGUUCGCCUGCAGGC-GAAGGCUCCUGGCCAGGCACAUCCACAUACAUAUCCACAUCCACAUGCA---- ---(((((((.....))...(((...(((.((((((((.....))))))))-...))).....))).))))).............................---- ( -33.40, z-score = -1.34, R) >droEre2.scaffold_4845 17505074 96 + 22589142 ---GUGCCCCUGUCAUCAACGGUAAUGUCAGCCUGCAG-UCGCCUGCAGGC-GAAGGCUCCUGGCCAGGCACAUCCACAUCCACAUCCAUAUCUACAUCCA---- ---(((((.((((.....))))........((((((((-....))))))))-...(((.....))).))))).............................---- ( -30.50, z-score = -2.13, R) >droAna3.scaffold_12916 10949226 98 + 16180835 GUGCCACUG-UGUCCUGAUUGGAAAUGUCAGCCUGCAGUUUGCCUGCAGGCUACCAGC----CUUCGGGCC--GUCACCGGGCCACACCCCCUCGUCCUGCCGCA (((((.(.(-((.(((((..((...((..(((((((((.....)))))))))..)).)----).)))))..--..))).))).)))................... ( -36.20, z-score = -1.07, R) >dp4.chr4_group4 274959 81 + 6586962 ---GUGCCU-UGGCCUCAGUGG-AAUGUCGGCCUGCAGGCAGACUGCAGACUGCAGGCGAGGUUUCAGGCCCAUUCUCCGGACGAA------------------- ---......-.(((((....((-(((.(((.(((((((((.....))...)))))))))).))))))))))...............------------------- ( -30.20, z-score = 0.06, R) >droPer1.super_16 584497 74 - 1990440 ---GUGCCU-UGGCCUCAGUGG-AAUGUCGGCCUGCAG-------GCAGACUGCAGGCGAGGUUUCAGGCCCAUUCUCCGGACGAA------------------- ---......-.(((((....((-(((.(((.(((((((-------.....)))))))))).))))))))))...............------------------- ( -30.60, z-score = -0.95, R) >consensus ___GUGCCGCUGUCAUCAUCGGUAAUGUCAGCCUGCAGUUCGCCUGCAGGC_GAAGGCUCCUGGCCAGGCACAUCCACAUGCACAUCCACAUCCA__________ .....(((..((....))..(((...(((.((((((((.....))))))))....))).....))).)))................................... (-16.02 = -16.70 + 0.68)

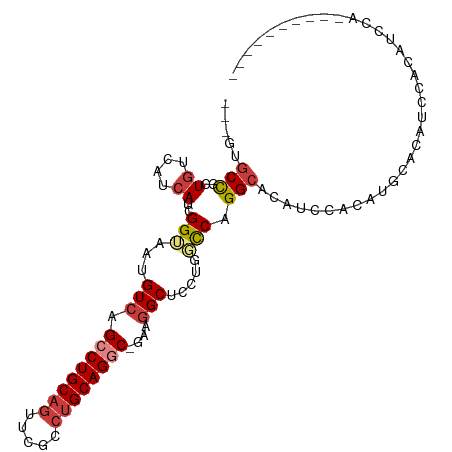
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:30 2011