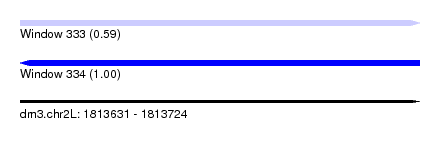
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,813,631 – 1,813,724 |
| Length | 93 |
| Max. P | 0.996753 |

| Location | 1,813,631 – 1,813,724 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.03 |
| Shannon entropy | 0.48461 |
| G+C content | 0.40736 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.67 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
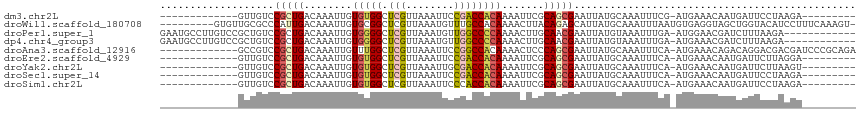
>dm3.chr2L 1813631 93 + 23011544 -------------GUUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCG-AUGAAACAAUGAUUCCUAAGA--------- -------------.......((((.(.((((.(((((.(((........)))))))).)))).)))))(((((((.....((((.-..))))..)))))))......--------- ( -21.00, z-score = -1.41, R) >droWil1.scaffold_180708 696023 106 - 12563649 ---------GUGUUGCGCCCAUUGACAAAUUGUGCGGCUCGUUAAAUGUUUGCCACAAAACUUACAGAGCAUUAUGCAAAUUUAAUGUGAGGUAGCUGGUACAUCCUUUCAAAGU- ---------(.(((((((..(((....))).))))))).).....((((..((((.....((((((..((.....))........)))))).....))))))))...........- ( -19.00, z-score = 1.81, R) >droPer1.super_1 336384 103 - 10282868 GAAUGCCUUGUCCGCUGUCCGCUGACAAAUUGUGGGGCUCGUUAAAUGUUGGCCCCAAAACUUGCAACGAAUUAUGUAAAUUUGA-AUGGAACGAUCUUUAAGA------------ ...(((.(((((.((.....)).)))))....((((((.((........))))))))......))).((((((.....)))))).-..................------------ ( -23.50, z-score = -0.70, R) >dp4.chr4_group3 3195376 103 - 11692001 GAAUGCCUUGUCCGCUGUCCGCUGACAAAUUGUGGGGCUCGUUAAAUGUUGGCCCCAAAACUUGCAACGAAUUAUGUAAAUUUGA-AUGAAACGAUCUUUAAGA------------ ...(((.(((((.((.....)).)))))....((((((.((........))))))))......))).((((((.....)))))).-..................------------ ( -23.50, z-score = -1.20, R) >droAna3.scaffold_12916 406505 102 - 16180835 -------------GCCGUCCGCUGACAAAUUGUUUGGCUCGUUAAAUUCCGGCCACAAAACUCCCAGCGAAUUAUGCAAAUUUCA-AUGAAACAGACAGGACGACGAUCCCGCAGA -------------..(((((.(((....((((..((((.((........))))))...........(((.....)))......))-))....)))...)))))............. ( -20.90, z-score = -0.02, R) >droEre2.scaffold_4929 1858584 93 + 26641161 -------------GUUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCA-AUGAAACAAUGAUUCUUAGGA--------- -------------....(((((((.(.((((.(((((.(((........)))))))).)))).)))))(((((((.....((((.-..))))..)))))))...)))--------- ( -24.10, z-score = -2.24, R) >droYak2.chr2L 1786652 93 + 22324452 -------------GUUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUGCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCA-AUGAAACAAUGAUUCUUAAGU--------- -------------.......((((.(.((((.(((((.((((......))))))))).)))).)))))(((((((.....((((.-..))))..)))))))......--------- ( -23.90, z-score = -2.07, R) >droSec1.super_14 364345 93 - 2068291 -------------GUUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCA-AUGAAACAAUGAUUCCUAAGA--------- -------------.......((((.(.((((.(((((.(((........)))))))).)))).)))))(((((((.....((((.-..))))..)))))))......--------- ( -21.00, z-score = -1.75, R) >droSim1.chr2L 1797001 93 + 22036055 -------------GUUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCCACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCA-AUGAAACAAUGAUUCCUAAGA--------- -------------.......((((.(.((((.(((((...............))))).)))).)))))(((((((.....((((.-..))))..)))))))......--------- ( -16.56, z-score = -0.53, R) >consensus _____________GUUGUCCGCUGACAAAUUGUGUGGCUCGUUAAAUUCCGACCACAAAAUUCGCAGCGAAUUAUGCAAAUUUCA_AUGAAACAAUGAUUCCUAAGA_________ ...................(((((........(((((.(((........)))))))).......)))))............................................... (-13.73 = -13.67 + -0.06)

| Location | 1,813,631 – 1,813,724 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.03 |
| Shannon entropy | 0.48461 |
| G+C content | 0.40736 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -21.62 |
| Energy contribution | -20.71 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
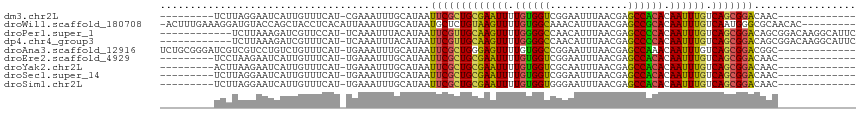
>dm3.chr2L 1813631 93 - 23011544 ---------UCUUAGGAAUCAUUGUUUCAU-CGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAAC------------- ---------.....(((((....)))))..-..............(((((((((((((.((((((((........))).))))).)))))).)))))))....------------- ( -27.70, z-score = -3.24, R) >droWil1.scaffold_180708 696023 106 + 12563649 -ACUUUGAAAGGAUGUACCAGCUACCUCACAUUAAAUUUGCAUAAUGCUCUGUAAGUUUUGUGGCAAACAUUUAACGAGCCGCACAAUUUGUCAAUGGGCGCAACAC--------- -....((..(((..((....))..)))..))......((((.....((((((((((((.((((((.............)))))).)))))).))..))))))))...--------- ( -19.72, z-score = 0.89, R) >droPer1.super_1 336384 103 + 10282868 ------------UCUUAAAGAUCGUUCCAU-UCAAAUUUACAUAAUUCGUUGCAAGUUUUGGGGCCAACAUUUAACGAGCCCCACAAUUUGUCAGCGGACAGCGGACAAGGCAUUC ------------.......(.(((((((..-...((((.....)))).((((((((((.(((((((..........).)))))).)))))).)))))))..)))).)......... ( -25.40, z-score = -2.30, R) >dp4.chr4_group3 3195376 103 + 11692001 ------------UCUUAAAGAUCGUUUCAU-UCAAAUUUACAUAAUUCGUUGCAAGUUUUGGGGCCAACAUUUAACGAGCCCCACAAUUUGUCAGCGGACAGCGGACAAGGCAUUC ------------..................-..............(((((((((((((.(((((((..........).)))))).)))))).)))))))..((.......)).... ( -25.40, z-score = -2.52, R) >droAna3.scaffold_12916 406505 102 + 16180835 UCUGCGGGAUCGUCGUCCUGUCUGUUUCAU-UGAAAUUUGCAUAAUUCGCUGGGAGUUUUGUGGCCGGAAUUUAACGAGCCAAACAAUUUGUCAGCGGACGGC------------- ...........((((((((((..(((((..-.)))))..)))......((((((((((...((((((........)).))))...))))).))))))))))))------------- ( -31.80, z-score = -1.38, R) >droEre2.scaffold_4929 1858584 93 - 26641161 ---------UCCUAAGAAUCAUUGUUUCAU-UGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAAC------------- ---------(((...((((...(((.....-........)))..))))((((((((((.((((((((........))).))))).)))))).)))))))....------------- ( -27.32, z-score = -3.21, R) >droYak2.chr2L 1786652 93 - 22324452 ---------ACUUAAGAAUCAUUGUUUCAU-UGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGCAAUUUAACGAGCCACACAAUUUGUCAGCGGACAAC------------- ---------......(((.......)))..-..............(((((((((((((.((((((((........))).))))).)))))).)))))))....------------- ( -25.40, z-score = -2.71, R) >droSec1.super_14 364345 93 + 2068291 ---------UCUUAGGAAUCAUUGUUUCAU-UGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAAC------------- ---------.....(((((....)))))..-..............(((((((((((((.((((((((........))).))))).)))))).)))))))....------------- ( -27.70, z-score = -3.29, R) >droSim1.chr2L 1797001 93 - 22036055 ---------UCUUAGGAAUCAUUGUUUCAU-UGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUGGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAAC------------- ---------.....(((((....)))))..-..............(((((((((((((.((((((.(........)..)))))).)))))).)))))))....------------- ( -25.40, z-score = -2.41, R) >consensus _________UCUUAGGAAUCAUUGUUUCAU_UGAAAUUUGCAUAAUUCGCUGCGAAUUUUGUGGUCGGAAUUUAACGAGCCACACAAUUUGUCAGCGGACAAC_____________ .............................................(((((((((((((.((((((.............)))))).)))))).)))))))................. (-21.62 = -20.71 + -0.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:17 2011