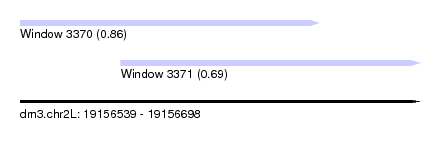
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,156,539 – 19,156,698 |
| Length | 159 |
| Max. P | 0.864465 |

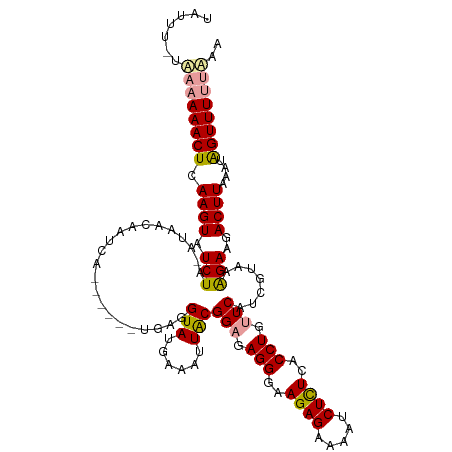
| Location | 19,156,539 – 19,156,658 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Shannon entropy | 0.29673 |
| G+C content | 0.30170 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -17.77 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19156539 119 + 23011544 UAUUUAUGAAAAACUCAAGUAUCUUCAUUUAAGUUAACUAACUGAGGUAUGAAAAUGCGGUGAGGGAAGAGAAAAUCUUUCACCUGUUCUUUCUAAGGAAGACUUAAAUGGUUUUUUAA ......((((((((..(((...)))((((((((((..((....(((((((....))))(((((((((........)))))))))....)))....))...)))))))))))))))))). ( -29.50, z-score = -2.46, R) >droSim1.chr2L 18846182 105 + 22036055 -------UAUAAACUCAAGUAUCUA-AUAACAAUCG------UGAGGUAUGAAAUUACGGAGAGGGAAGAGAAAAUCUCUCACCUGUUCAUCGUAAAGAAGACUUAAAUAGUUUUUAAA -------...........((((((.-...((....)------).))))))....(((((((.(((..((((.....))))..))).))...))))).(((((((.....)))))))... ( -22.10, z-score = -1.67, R) >droSec1.super_7 2778972 111 + 3727775 UAUUUUUAAAAAACUCAAGUAUCUA-AUAACAAUCA------UGAUGUAUGAAAUUACGGAGAGGGAAGAGAAAAUCUCUCACCUGUUCAUCGCAAAGAAGACUUAAAUAGUUUUAAA- .........((((((.((((.(((.-.......(((------((...)))))......(((.(((..((((.....))))..))).))).......)))..))))....))))))...- ( -20.30, z-score = -1.47, R) >consensus UAUUU_UAAAAAACUCAAGUAUCUA_AUAACAAUCA______UGAGGUAUGAAAUUACGGAGAGGGAAGAGAAAAUCUCUCACCUGUUCAUCGUAAAGAAGACUUAAAUAGUUUUUAAA .......((((((((.((((.(((......................(((......)))(((.(((..((((.....))))..))).))).......)))..))))....)))))))).. (-17.77 = -18.00 + 0.23)


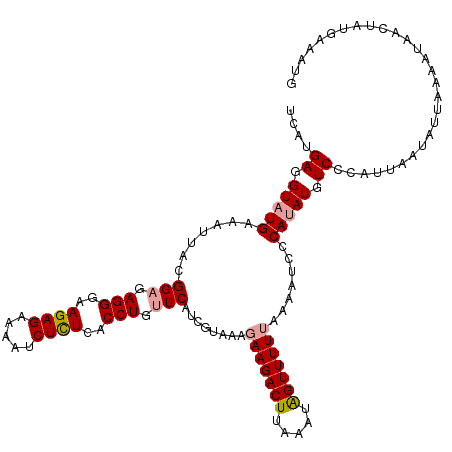
| Location | 19,156,579 – 19,156,698 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Shannon entropy | 0.16626 |
| G+C content | 0.31842 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19156579 119 + 23011544 -ACUGAGGUAUGAAAAUGCGGUGAGGGAAGAGAAAAUCUUUCACCUGUUCUUUCUAAGGAAGACUUAAAUGGUUUUUUAAAUCCCACAUGUCGCAUUAAUAUAAAAAUAACUAUGAAAUG -......((((...(((((((((.((((..((((((((........((.((((.....))))))......))))))))...)))).)))..)))))).)))).................. ( -23.74, z-score = -1.02, R) >droSim1.chr2L 18846207 120 + 22036055 UCGUGAGGUAUGAAAUUACGGAGAGGGAAGAGAAAAUCUCUCACCUGUUCAUCGUAAAGAAGACUUAAAUAGUUUUUAAAACCCCAUAUGUCCCAUUAAUAUUAAAAUAACUAUGAAAUG ..(.((.(((((...(((((((.(((..((((.....))))..))).))...))))).(((((((.....))))))).......))))).)).).......................... ( -25.30, z-score = -1.99, R) >droSec1.super_7 2779004 119 + 3727775 UCAUGAUGUAUGAAAUUACGGAGAGGGAAGAGAAAAUCUCUCACCUGUUCAUCGCAAAGAAGACUUAAAUAGUUUU-AAAAUCCCAUAUAUCCCAUUAAUAUUAAAAUAACUAUGAAAUG ....((((((((.......(((.(((..((((.....))))..))).))).........((((((.....))))))-.......))))))))............................ ( -24.90, z-score = -2.68, R) >consensus UCAUGAGGUAUGAAAUUACGGAGAGGGAAGAGAAAAUCUCUCACCUGUUCAUCGUAAAGAAGACUUAAAUAGUUUUUAAAAUCCCAUAUGUCCCAUUAAUAUUAAAAUAACUAUGAAAUG ....((.(((((.......(((.(((..((((.....))))..))).)))........(((((((.....))))))).......))))).))............................ (-19.54 = -20.10 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:27 2011