| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,143,153 – 19,143,263 |
| Length | 110 |
| Max. P | 0.545232 |

| Location | 19,143,153 – 19,143,263 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.74 |
| Shannon entropy | 0.67785 |
| G+C content | 0.47516 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -13.38 |
| Energy contribution | -12.67 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
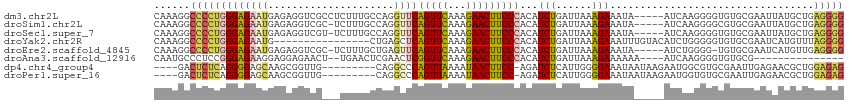
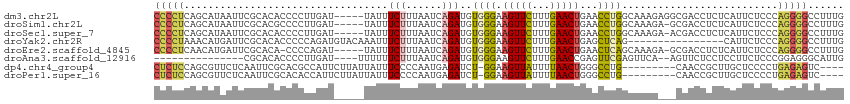
>dm3.chr2L 19143153 110 + 23011544 CAAAGGCCCCUGGGAGAAUGAGAGGUCGCCUCUUUGCCAGGUUCAGUUCAAAGAACUUCCCACAUCUGAUUAAAGAAAUA-----AUCAAGGGGUGUGCGAAUUAUGCUGAGGGG ......(((((.((...((((...((((((((((((..((((((........))))))..)).....(((((......))-----))))))))))).))...)))).)).))))) ( -31.40, z-score = -0.23, R) >droSim1.chr2L 18840032 109 + 22036055 CAAAGGCCCCUGGGAGAAUGAGAGGUCGC-UCUUUGCCAGGUUCAGUUCAAAGAACUUCCCACAUCUGAUUAAAGAAAUA-----AUCAAGGGGCGUGCGAAUUAUGCUGAGGGG ......(((((.((...((((....((((-....((((.((...(((((...)))))..)).....((((((......))-----))))...)))).)))).)))).)).))))) ( -32.40, z-score = -0.65, R) >droSec1.super_7 2772681 109 + 3727775 CAAAGGCCCCUGGGAGAAUGAGAGGUCGU-UCUUUGCCAGGUUCAGUUCAAAGAACUUCCCACAUCUGAUUAAAGAAAUA-----AUCAAGGGGUGUGCGAAUUAUGCUGAGGGG ......(((((.(((((((((....))))-)))))..(((((((........))))(((.((((((((((((......))-----)))...))))))).))).....)))))))) ( -32.20, z-score = -0.94, R) >droYak2.chr2R 5658184 99 + 21139217 CAAAGGCCCCUGGGAGAAUG----------------CUGAGCUCAGUUCAAAGAACUUCCCACAUCUGAUUAAAGAAAUUUGUACAUCUGGGGGUGUGCGAAUCAUGUUUAGGGG ......((((((((((....----------------.(((((...)))))......)))))((((((......))).(((((((((((....)))))))))))..)))..))))) ( -29.30, z-score = -1.02, R) >droEre2.scaffold_4845 17459223 108 - 22589142 CAAAGGCCCCUGGGAGAAUGAGAGGUCGC-UCUUUGCUGAGUUCAGUUCAAAGAACUUCCCACAUCUGAUUAAAGAAAUA-----AUCUGGGG-UGUGCGAAUCAUGUUGAGGGG ......(((((.((...((((....((((-(((((((((....)))..))))))((.(((((.....(((((......))-----))))))))-.)))))).)))).)).))))) ( -33.70, z-score = -1.29, R) >droAna3.scaffold_12916 10896218 94 - 16180835 CAAUGCCCUCCGGGAGAAGGAGGAGAACU--UGAACUCGAACUCGGUUCAAAGAACUUCCCACAUCUGAUUAAAGAAAAAA----AUCAAGGGGUGUGCG--------------- ....(((((((..((...((.((((..((--(((((.((....)))))).)))..))))))...(((......))).....----.))..)))).).)).--------------- ( -24.50, z-score = -0.74, R) >dp4.chr4_group4 224740 101 - 6586962 ----GACUCUCAGGGGAGCAAGCGGUUG---------CAGGCCCAGUUAAAAUAACUUCC-AGAUCUCAUUGGGGAAAUAAUAAGAAUGGCGUGCGAAUUGAGAACGCUGGAGAG ----..(((((((.(...(((....(((---------((.(((.(((((...)))))(((-((......)))))..............))).))))).)))....).)).))))) ( -26.30, z-score = -0.45, R) >droPer1.super_16 530629 101 + 1990440 ----GACUCUCAGGGGAGCAAGCGGUUG---------CAGGCCCAGUUAAAAUAACUUCC-AGAUCUCAUUGGGGAAAUAAUAAGAAUGGUGUGCGAAUUGAGAACGCUGGAGAG ----..(((((((.(...(((....(((---------((.(((.(((((...)))))(((-((......)))))..............))).))))).)))....).)).))))) ( -24.80, z-score = -0.11, R) >consensus CAAAGGCCCCUGGGAGAAUGAGAGGUCG___CUUUGCCAGGCUCAGUUCAAAGAACUUCCCACAUCUGAUUAAAGAAAUA_____AUCAAGGGGUGUGCGAAUUAUGCUGAGGGG ......(((((((((((((.....................))))(((((...)))))))))...(((......)))..................................))))) (-13.38 = -12.67 + -0.70)
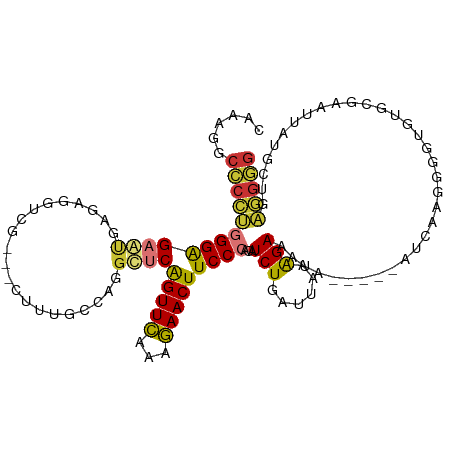
| Location | 19,143,153 – 19,143,263 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 65.74 |
| Shannon entropy | 0.67785 |
| G+C content | 0.47516 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -9.56 |
| Energy contribution | -8.82 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19143153 110 - 23011544 CCCCUCAGCAUAAUUCGCACACCCCUUGAU-----UAUUUCUUUAAUCAGAUGUGGGAAGUUCUUUGAACUGAACCUGGCAAAGAGGCGACCUCUCAUUCUCCCAGGGGCCUUUG (((((..((....(((.((((....(((((-----((......))))))).)))).)))((((........))))...))..(((((...))))).........)))))...... ( -28.50, z-score = -0.80, R) >droSim1.chr2L 18840032 109 - 22036055 CCCCUCAGCAUAAUUCGCACGCCCCUUGAU-----UAUUUCUUUAAUCAGAUGUGGGAAGUUCUUUGAACUGAACCUGGCAAAGA-GCGACCUCUCAUUCUCCCAGGGGCCUUUG .......((.......))..((((((((((-----((......)))))))...(((((.((((((((............))))))-))((....))....))))))))))..... ( -31.00, z-score = -1.79, R) >droSec1.super_7 2772681 109 - 3727775 CCCCUCAGCAUAAUUCGCACACCCCUUGAU-----UAUUUCUUUAAUCAGAUGUGGGAAGUUCUUUGAACUGAACCUGGCAAAGA-ACGACCUCUCAUUCUCCCAGGGGCCUUUG .......((.......))...(((((((((-----((......)))))))...(((((.((((((((............))))))-))((....))....)))))))))...... ( -27.40, z-score = -1.39, R) >droYak2.chr2R 5658184 99 - 21139217 CCCCUAAACAUGAUUCGCACACCCCCAGAUGUACAAAUUUCUUUAAUCAGAUGUGGGAAGUUCUUUGAACUGAGCUCAG----------------CAUUCUCCCAGGGGCCUUUG (((((......((...((...(((.((..((...(((....)))...))..)).))).(((((........)))))..)----------------)..))....)))))...... ( -19.70, z-score = 0.44, R) >droEre2.scaffold_4845 17459223 108 + 22589142 CCCCUCAACAUGAUUCGCACA-CCCCAGAU-----UAUUUCUUUAAUCAGAUGUGGGAAGUUCUUUGAACUGAACUCAGCAAAGA-GCGACCUCUCAUUCUCCCAGGGGCCUUUG (((((....(((((((.((((-.(...(((-----((......))))).).)))).)))((((((((..(((....)))))))))-))......))))......)))))...... ( -29.60, z-score = -2.08, R) >droAna3.scaffold_12916 10896218 94 + 16180835 ---------------CGCACACCCCUUGAU----UUUUUUCUUUAAUCAGAUGUGGGAAGUUCUUUGAACCGAGUUCGAGUUCA--AGUUCUCCUCCUUCUCCCGGAGGGCAUUG ---------------.((...(((((((((----(.........))))))..).)))......(((((((((....)).)))))--))....(((((.......))))))).... ( -23.70, z-score = -0.67, R) >dp4.chr4_group4 224740 101 + 6586962 CUCUCCAGCGUUCUCAAUUCGCACGCCAUUCUUAUUAUUUCCCCAAUGAGAUCU-GGAAGUUAUUUUAACUGGGCCUG---------CAACCGCUUGCUCCCCUGAGAGUC---- (((((..(((.........)))...(((.(((((((........)))))))..)-)).(((((...)))))(((...(---------(((....))))..))).)))))..---- ( -22.80, z-score = -1.21, R) >droPer1.super_16 530629 101 - 1990440 CUCUCCAGCGUUCUCAAUUCGCACACCAUUCUUAUUAUUUCCCCAAUGAGAUCU-GGAAGUUAUUUUAACUGGGCCUG---------CAACCGCUUGCUCCCCUGAGAGUC---- (((((..(((.........)))...(((.(((((((........)))))))..)-)).(((((...)))))(((...(---------(((....))))..))).)))))..---- ( -23.10, z-score = -1.69, R) >consensus CCCCUCAGCAUAAUUCGCACACCCCCUGAU_____UAUUUCUUUAAUCAGAUGUGGGAAGUUCUUUGAACUGAACCUGGCAAAG___CGACCUCUCAUUCUCCCAGGGGCCUUUG (((((..................................(((......)))..((((.(((((...)))))...))))..........................)))))...... ( -9.56 = -8.82 + -0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:25 2011