| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,122,142 – 19,122,245 |
| Length | 103 |
| Max. P | 0.999067 |

| Location | 19,122,142 – 19,122,245 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 53.09 |
| Shannon entropy | 0.90496 |
| G+C content | 0.49184 |
| Mean single sequence MFE | -29.09 |
| Consensus MFE | -7.48 |
| Energy contribution | -7.51 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973827 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
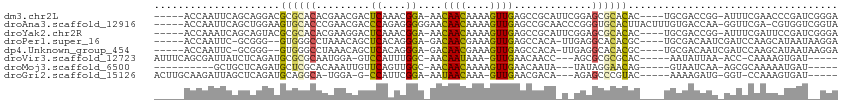
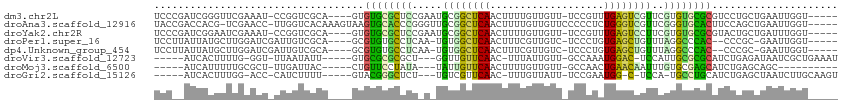
>dm3.chr2L 19122142 103 + 23011544 -----ACCAAUUCAGCAGGACGCGCACACGAACGACUCAAACGGA-AACAACAAAAGUUGAGCCGCAUUCGGAGCGCACAC----UGCGACCGG-AUUUCGAACCCGAUCGGGA -----.((......((((...((((.(.((((((.(((....(..-..)(((....)))))).))..))))).))))...)----)))((.(((-.........))).)))).. ( -30.80, z-score = -2.06, R) >droAna3.scaffold_12916 10876995 107 - 16180835 -----ACCAAUUCAGCUGGAAGUGCACCCGAACGACCCAGAGGGGGAACAACAAAAGUUGAGCCGCAACCCGGGUGCACUUACUUUGUGACCAA-GGUUCGA-CGUGGUCGGUA -----(((......((.(((((((((((((.....(((...)))((..((((....))))..))......))))))))))).))..))(((((.-(......-).)))))))). ( -41.90, z-score = -2.40, R) >droYak2.chr2R 5637778 103 + 21139217 -----ACCAAAUCAGCAGUACGCGCACACGAAGGACUCAAACGGA-AACAACAAAAGUUGAGCCGCAUUCGGAGCGCACAC----UGCGACCGG-AUUUCGAUUCCGAUCGGGA -----.((......(((((..((((.(.((((((.(((....(..-..)(((....))))))))...))))).))))..))----)))((.(((-(.......)))).)))).. ( -34.50, z-score = -3.15, R) >droPer1.super_16 511419 100 + 1990440 -----ACCAAUUC-GCGGG--GUGGGCCUAAACAGCUCACAGGGA-GACAACGAAAGUUGAGCCACA-UUGAGGCACACGC----UGCGACAAUCGAUCCAAGCAUAAUAAGGA -----......((-((((.--(((.((((.....((((....(..-..)(((....)))))))....-...)))).))).)----))))).......(((...........))) ( -33.10, z-score = -3.24, R) >dp4.Unknown_group_454 11527 100 + 26276 -----ACCAAUUC-GCGGG--GUGGGCCUAAACAGCUCACAGGGA-GACAACGAAAGUUGAGCCACA-UUGAGGCACACGC----UGCGACAAUCGAUCCAAGCAUAAUAAGGA -----......((-((((.--(((.((((.....((((....(..-..)(((....)))))))....-...)))).))).)----))))).......(((...........))) ( -33.10, z-score = -3.24, R) >droVir3.scaffold_12723 3076833 96 + 5802038 AUUUCAGCGAUUAUCUCAGAUGCGCGCAAUGGA-GUCCAUUUGGC-AACAAUAAA-GUUGAACAACC---AGCGCGCGCAC-----AAUAUUAA-ACC-CAAAAGUGAU----- ....................(((((((..(((.-..((....))(-(((......-)))).....))---)..))))))).-----........-...-..........----- ( -21.30, z-score = -0.87, R) >droMoj3.scaffold_6500 25570539 89 - 32352404 ----------GCUGCUCAGAUGCUCGCACAAAUUGUUCAGUUGGC-AACAACAAAAGUUGAACAAUA---UAUAGGAACAG-----GUAAUCAA-AGCGCAAAAAUGAU----- ----------((.(((..((((((..(....((((((((((((..-..))))......)))))))).---.....)....)-----)).)))..-))))).........----- ( -18.50, z-score = -1.08, R) >droGri2.scaffold_15126 7993723 94 - 8399593 ACUUGCAAGAUUAGCUCAGAUGCAGGCA-UGGA-G-CCAUUCGGA-AAUAACAAA-GUUGAACGACA---AGAGCCCGUAC-----AAAAGAUG-GGU-CCAAAGUGAU----- ..(..(..((...((((..(((....))-).))-)-)...))(((-.........-(((....))).---....(((((..-----.....)))-)))-))...)..).----- ( -19.50, z-score = -0.28, R) >consensus _____ACCAAUUCAGCCGGAUGCGCGCACAAACGGCUCAAACGGA_AACAACAAAAGUUGAGCCACA_U_GGAGCGCACAC____UGCGACCAA_AGUUCAAACAUGAU__GGA .....................((((((.........((....))....((((....)))).............))))))................................... ( -7.48 = -7.51 + 0.03)
| Location | 19,122,142 – 19,122,245 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 53.09 |
| Shannon entropy | 0.90496 |
| G+C content | 0.49184 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -14.73 |
| Energy contribution | -13.37 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 2.35 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999067 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
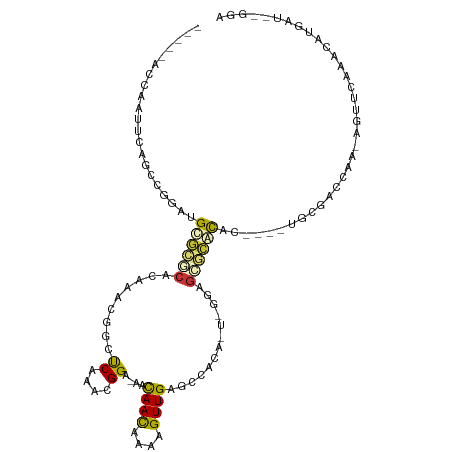
>dm3.chr2L 19122142 103 - 23011544 UCCCGAUCGGGUUCGAAAU-CCGGUCGCA----GUGUGCGCUCCGAAUGCGGCUCAACUUUUGUUGUU-UCCGUUUGAGUCGUUCGUGUGCGCGUCCUGCUGAAUUGGU----- ..((((((((((.....))-))))))(((----(..(((((.((((..(((((((((...........-.....)))))))))))).).)))))..))))......)).----- ( -42.49, z-score = -3.90, R) >droAna3.scaffold_12916 10876995 107 + 16180835 UACCGACCACG-UCGAACC-UUGGUCACAAAGUAAGUGCACCCGGGUUGCGGCUCAACUUUUGUUGUUCCCCCUCUGGGUCGUUCGGGUGCACUUCCAGCUGAAUUGGU----- .((((((((.(-......)-.)))))....((((((((((((((..(.((((..((((....))))..))((....)))).)..)))))))))))...))).....)))----- ( -40.50, z-score = -2.23, R) >droYak2.chr2R 5637778 103 - 21139217 UCCCGAUCGGAAUCGAAAU-CCGGUCGCA----GUGUGCGCUCCGAAUGCGGCUCAACUUUUGUUGUU-UCCGUUUGAGUCCUUCGUGUGCGCGUACUGCUGAUUUGGU----- ..(((((((((.......)-))))))(((----((((((((.(((((.(.(((((((...........-.....)))))))))))).).)))))))))))......)).----- ( -40.69, z-score = -3.75, R) >droPer1.super_16 511419 100 - 1990440 UCCUUAUUAUGCUUGGAUCGAUUGUCGCA----GCGUGUGCCUCAA-UGUGGCUCAACUUUCGUUGUC-UCCCUGUGAGCUGUUUAGGCCCAC--CCCGC-GAAUUGGU----- (((...........)))(((((..((((.----(.(((.((((...-.(..(((((......(....)-......)))))..)..)))).)))--.).))-))))))).----- ( -26.00, z-score = -0.53, R) >dp4.Unknown_group_454 11527 100 - 26276 UCCUUAUUAUGCUUGGAUCGAUUGUCGCA----GCGUGUGCCUCAA-UGUGGCUCAACUUUCGUUGUC-UCCCUGUGAGCUGUUUAGGCCCAC--CCCGC-GAAUUGGU----- (((...........)))(((((..((((.----(.(((.((((...-.(..(((((......(....)-......)))))..)..)))).)))--.).))-))))))).----- ( -26.00, z-score = -0.53, R) >droVir3.scaffold_12723 3076833 96 - 5802038 -----AUCACUUUUG-GGU-UUAAUAUU-----GUGCGCGCGCU---GGUUGUUCAAC-UUUAUUGUU-GCCAAAUGGAC-UCCAUUGCGCGCAUCUGAGAUAAUCGCUGAAAU -----......(((.-(((-....((((-----(((((((((.(---((..((.((((-......)))-)((....))))-.))).)))))))))....))))...))).))). ( -25.20, z-score = -0.82, R) >droMoj3.scaffold_6500 25570539 89 + 32352404 -----AUCAUUUUUGCGCU-UUGAUUAC-----CUGUUCCUAUA---UAUUGUUCAACUUUUGUUGUU-GCCAACUGAACAAUUUGUGCGAGCAUCUGAGCAGC---------- -----...........(((-(.((....-----.(((((.((((---.((((((((......((((..-..)))))))))))).)))).))))))).))))...---------- ( -20.90, z-score = -1.99, R) >droGri2.scaffold_15126 7993723 94 + 8399593 -----AUCACUUUGG-ACC-CAUCUUUU-----GUACGGGCUCU---UGUCGUUCAAC-UUUGUUAUU-UCCGAAUGG-C-UCCA-UGCCUGCAUCUGAGCUAAUCUUGCAAGU -----...((((.((-(..-...(((..-----((.(((((...---.(((((((...-.........-...))))))-)-....-.))))).))..)))....)))...)))) ( -17.96, z-score = 0.08, R) >consensus UCC__AUCACGUUUGAACC_CUGGUCGCA____GUGUGCGCCCC_A_UGUGGCUCAACUUUUGUUGUU_UCCCUAUGAGCCGUUCGUGCGCGCAUCCGGCUGAAUUGGU_____ ..................................(((((((((.....((((((((...................))))))))..))))))))).................... (-14.73 = -13.37 + -1.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:22 2011