| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,103,639 – 19,103,736 |
| Length | 97 |
| Max. P | 0.959874 |

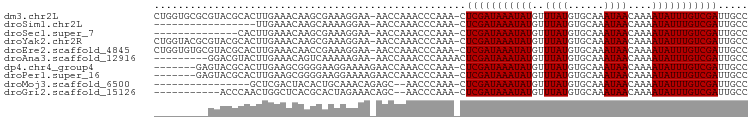
| Location | 19,103,639 – 19,103,736 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Shannon entropy | 0.37834 |
| G+C content | 0.36628 |
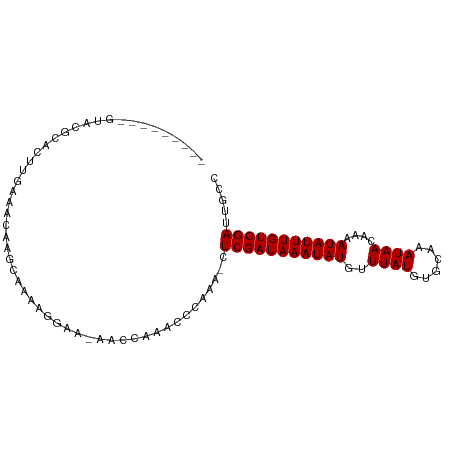
| Mean single sequence MFE | -17.94 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19103639 97 + 23011544 CUGGUGCGCGUACGCACUUGAAACAAGCGAAAGGAA-AACCAAACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC (.((((((....)))))).)......((((..((..-..)).........-.(((((((((((..((((......))))....))))))))))))))). ( -22.60, z-score = -2.34, R) >droSim1.chr2L 18800322 80 + 22036055 -----------------UUGAAACAAGCAAAAGGAA-AACCAAACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC -----------------.........((((..((..-..)).........-.(((((((((((..((((......))))....))))))))))))))). ( -15.20, z-score = -3.09, R) >droSec1.super_7 2733217 83 + 3727775 --------------CACUUGAAACAAGCGAAAGGAA-AACCAAACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC --------------............((((..((..-..)).........-.(((((((((((..((((......))))....))))))))))))))). ( -14.90, z-score = -2.64, R) >droYak2.chr2R 5619336 97 + 21139217 CUGGUACGCGUACGCACUUGAAACAAGCGAAAGGAA-AACCAAACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC .((((..((....))............(....)...-.))))........-.(((((((((((..((((......))))....)))))))))))..... ( -17.90, z-score = -1.06, R) >droEre2.scaffold_4845 17420680 97 - 22589142 CUGGUGUGCGUACGCACUUGAAACAACCGAAAGGAA-AACCAAACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC .((((((((....)))).........((....))..-.))))........-.(((((((((((..((((......))))....)))))))))))..... ( -24.80, z-score = -3.56, R) >droAna3.scaffold_12916 10858827 89 - 16180835 ---------GGACGUACUUGAAACAGUCAAAAAGAA-AACCAAACCCAAAACUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC ---------.(((............)))........-...............(((((((((((..((((......))))....)))))))))))..... ( -12.90, z-score = -1.31, R) >dp4.chr4_group4 210485 91 - 6586962 -------GAGUACGCACUUGAAGCGGGGAAGGAAAAGAACCAAACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC -------((((.(((.......)))(((..((.......))...)))..)-)))(((((((((..((((......))))....)))))))))....... ( -20.10, z-score = -2.18, R) >droPer1.super_16 485715 91 + 1990440 -------GAGUACGCACUUGAAGCGGGGAAGGAAAAGAACCAAACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC -------((((.(((.......)))(((..((.......))...)))..)-)))(((((((((..((((......))))....)))))))))....... ( -20.10, z-score = -2.18, R) >droMoj3.scaffold_6500 25545669 80 - 32352404 ----------------GCUCGACUACACUGCAAACAGAGC--AACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC ----------------((((................))))--........-.(((((((((((..((((......))))....)))))))))))..... ( -15.89, z-score = -2.63, R) >droGri2.scaffold_15126 7973887 85 - 8399593 -----------ACCCAACUGGCUCACGCACUAGAAACAGC--AACCCAAA-CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC -----------......((((........)))).....((--((......-.(((((((((((..((((......))))....))))))))))))))). ( -15.01, z-score = -1.77, R) >consensus _________GUACGCACUUGAAACAAGCAAAAGGAA_AACCAAACCCAAA_CUCGAUAAAUAUGUUUAUGUGCAAAUAACAAAAUAUUUGUCGAUUGCC ....................................................(((((((((((..((((......))))....)))))))))))..... (-11.10 = -11.10 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:20 2011