| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,100,049 – 19,100,146 |
| Length | 97 |
| Max. P | 0.889457 |

| Location | 19,100,049 – 19,100,146 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 64.03 |
| Shannon entropy | 0.64161 |
| G+C content | 0.43597 |
| Mean single sequence MFE | -19.17 |
| Consensus MFE | -8.79 |
| Energy contribution | -8.61 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
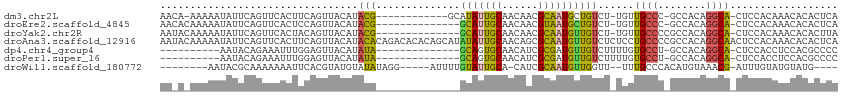
>dm3.chr2L 19100049 97 + 23011544 AACA-AAAAAUAUUCAGUUCACUUCAGUUACAUACG------------GCAUAUUGCAACAACGCAAUGCUGUCU-UGUUGCCC-GCCACAGGCA-CUCCACAAACACACUCA ....-...........(((.......((.(((.(((------------((((..(((......))))))))))..-))).))..-(((...))).-.......)))....... ( -16.70, z-score = -0.96, R) >droEre2.scaffold_4845 17417264 96 - 22589142 AACACAAAAAUAUUCAGUUCACUCCAGUUACAUACG--------------GCAUUGCAACAACGUAAUGCUGUCU-UGUUGCCC-GCCACAGGCA-CUCCACAAACACACUCA ................(((.......((.(((.(((--------------(((((((......))))))))))..-))).))..-(((...))).-.......)))....... ( -19.40, z-score = -2.01, R) >droYak2.chr2R 5616256 97 + 21139217 AAUACAAAAAUAUUCAGUUCACUACAGUUACAUACG--------------GCAUUGCAACAACGCAAUGUUGUCU-UGUUGCCCCGCCACAGGCA-CUCCACAAACACACUUA ..........................((((((.(((--------------(((((((......))))))))))..-)))((((........))))-.......)))....... ( -18.90, z-score = -1.64, R) >droAna3.scaffold_12916 10854997 113 - 16180835 AAUACAAAAAUAUUCAGUUCACUUCAGUUACAUACACAGACACACAGCAUAUAUUGCAACAGCGCAAUGUUGUCUCUCCUGCCCCGCCACAGGCAACUCCACAAACACACUCA ..........................(((........(((((........(((((((......))))))))))))....((((........))))........)))....... ( -14.50, z-score = -0.60, R) >dp4.chr4_group4 206876 87 - 6586962 ----------AAUACAGAAAUUUGGAGUUACAUAUA--------------GCAGUGCAACAUCGCGAUGUUGUCUUUUGUGCCU-GCCACAGGCA-CUCCACCUCCACGCCCC ----------............(((((......(((--------------(((.(((......))).)))))).....((((((-(...))))))-).....)))))...... ( -23.70, z-score = -2.15, R) >droPer1.super_16 482078 87 + 1990440 ----------AAUACAGAAAUUUGGAGUUACAUAUA--------------GCAGUGCAACAUCGCGAUGUUGUCUUUUGUGCCU-GCCACAGGCA-CUCCACCUCCACGCCCC ----------............(((((......(((--------------(((.(((......))).)))))).....((((((-(...))))))-).....)))))...... ( -23.70, z-score = -2.15, R) >droWil1.scaffold_180772 7065429 92 + 8906247 --------AAUACGCAAAAAAAUUCACGUAUGUAUAUAGG-----AUUUUGUAUUGCA-CAUCGCAAUGUUGGUU--UUUGCCCACAUGUAAACG-AUUUGUAUGUAUG---- --------.(((((............)))))(((((((.(-----(..((((..(((.-....)))((((.(((.--...))).))))....)))-).)).))))))).---- ( -17.30, z-score = 0.34, R) >consensus AA_A_AAAAAUAUUCAGUUCACUUCAGUUACAUACA______________GCAUUGCAACAACGCAAUGUUGUCU_UGUUGCCC_GCCACAGGCA_CUCCACAAACACACUCA ..................................................(((((((......))))))).........((((........)))).................. ( -8.79 = -8.61 + -0.18)
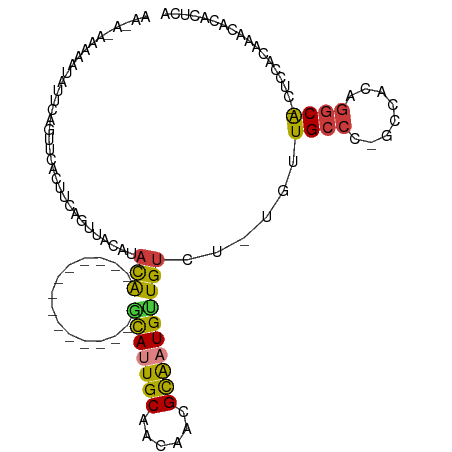

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:19 2011