| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,089,236 – 19,089,329 |
| Length | 93 |
| Max. P | 0.825164 |

| Location | 19,089,236 – 19,089,329 |
|---|---|
| Length | 93 |
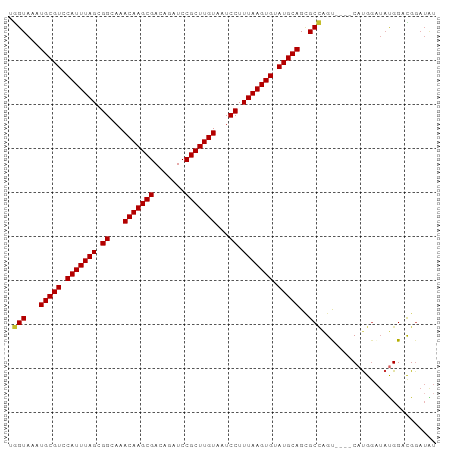
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Shannon entropy | 0.17099 |
| G+C content | 0.47734 |
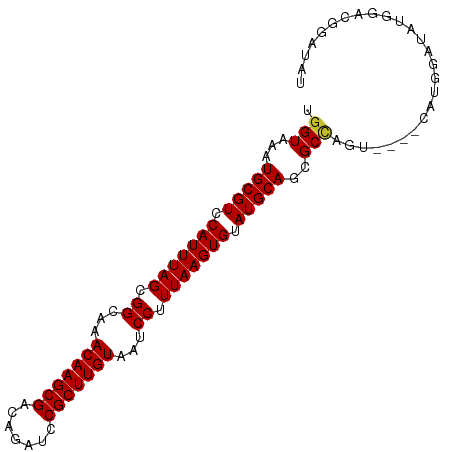
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -26.29 |
| Energy contribution | -26.31 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.825164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19089236 93 + 23011544 UGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCCAGU----CAUGGAUAUGGUCGGAUAU ((((...(((((.(((((((.((...(((((((.......)))))))...)).))))))).)))))..))))((----(.(((......)))))).. ( -28.30, z-score = -1.64, R) >droSim1.chr2L 18783032 93 + 22036055 UGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCCAGU----CAUGGAUAUGGUCGGAUAU ((((...(((((.(((((((.((...(((((((.......)))))))...)).))))))).)))))..))))((----(.(((......)))))).. ( -28.30, z-score = -1.64, R) >droSec1.super_7 2718869 93 + 3727775 UGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCUAGU----CAUGGAUAUGGUCGGAUAU ((((...(((((.(((((((.((...(((((((.......)))))))...)).))))))).)))))..))))((----(.(((......)))))).. ( -26.00, z-score = -0.92, R) >droYak2.chr2R 5605035 93 + 21139217 UGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCCAGU----CAUGGAUAUGGGCGGAUGU .......(((((.(((((((.((...(((((((.......)))))))...)).))))))).))))).((((.((----(...)))...))))..... ( -29.30, z-score = -1.19, R) >droEre2.scaffold_4845 17406374 93 - 22589142 UGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCCAGU----CAUGGAUAUGGGCGGAUAU .......(((((.(((((((.((...(((((((.......)))))))...)).))))))).))))).((((.((----(...)))...))))..... ( -29.30, z-score = -1.48, R) >droAna3.scaffold_12916 10842414 97 - 16180835 UGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGUGCCAGCACAGCACGGAUCUGGAUGGAUAU (((((..(((((.(((((((.((...(((((((.......)))))))...)).))))))).))))).)))))...........((((....)))).. ( -29.60, z-score = -1.37, R) >dp4.chr4_group4 196093 93 - 6586962 GGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCCAGC----CAUGGAUACGGACGGAUGU ......((.(((((((((((.((...(((((((.......)))))))...)).))))))((((.((((....))----..)).))))))))).)).. ( -29.90, z-score = -1.41, R) >droPer1.super_16 471271 93 + 1990440 GGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCCAGC----CAUGGAUACGGACGGAUGU ......((.(((((((((((.((...(((((((.......)))))))...)).))))))((((.((((....))----..)).))))))))).)).. ( -29.90, z-score = -1.41, R) >droWil1.scaffold_180772 7048311 90 + 8906247 GAGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGAGC-GGCAU--CAUGCGAAUGGAUGC---- ........((((((((((.((((...(((((((.......)))))))...)).....(((.((((.....-.))))--)))))))))))))))---- ( -31.40, z-score = -2.87, R) >consensus UGGUAAAUGCGUCCAUUUAGCGGCAAACAAGCGACAGAUCCGCUUGUAAUCCUUUAAGUGUAUGCAGCGCCAGU____CAUGGAUAUGGACGGAUAU .(((...(((((.(((((((.((...(((((((.......)))))))...)).))))))).)))))..))).......................... (-26.29 = -26.31 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:18 2011