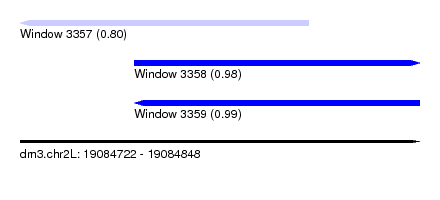
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,084,722 – 19,084,848 |
| Length | 126 |
| Max. P | 0.991362 |

| Location | 19,084,722 – 19,084,813 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Shannon entropy | 0.44474 |
| G+C content | 0.58328 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -14.64 |
| Energy contribution | -16.42 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19084722 91 - 23011544 ----------------------ACCGAAA--CCAAGAGGCCGAGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUCUCGUCUCGUCUCGCCUUACCAACCGACUGGCAAACU---- ----------------------.......--....(((((.((((.((..(((....((((........))))...))).)))))))))))(((.............))).....---- ( -25.02, z-score = -1.35, R) >droAna3.scaffold_12916 10838439 82 + 16180835 ----------------------GCCGAAA-CCAAAGAGACCAUGAGCGCCGACCACUUGGCAGCUCAUAAGCACACGUCUCGCCUCACCUCGCC------ACUUAUAGGCU-------- ----------------------(((....-.....(((((.(((((((((((....))))).))))))..(....))))))((........)).------.......))).-------- ( -24.10, z-score = -2.91, R) >droEre2.scaffold_4845 17402505 102 + 22589142 ------GCC--GAGACCAAAAGGCCGAGAGGCCGAGAGCCUGAGAGCGGCGACCACUUGGCAGCUCAUAGCCAUAAGUCUCGUCUCGCCUCACCU-----ACCGACUGGCAAACU---- ------(((--.((.......((((....))))(((.((..((((.(((.(((....((((........))))...)))))))))))))))....-----.....))))).....---- ( -37.00, z-score = -1.83, R) >droYak2.chr2R 5600536 119 - 21139217 GCCCAAACCAAGAGACCUAGAGACCGAGAGGUCGAGAGGCGAAGAGCAGCGACCACUUGGCAGCUCAUAGCCAUAAGUCUCGUCUCGCCUCACCUCACCAACCGACUGGCCAACAAACU ...........(((.......((((....))))..(((((((.((...(.(((....((((........))))...))).).)))))))))..))).(((......))).......... ( -35.90, z-score = -2.20, R) >droSec1.super_7 2714501 91 - 3727775 ----------------------GCCGAAA--CCAAGAGGCCGAGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUCUCGUCUCGUCUCGCCUCACCAACCGACUGGCAAACU---- ----------------------((((...--.)..((((((((((.((..(((....((((........))))...))).)))))))....)))))...........))).....---- ( -27.40, z-score = -1.76, R) >droSim1.chr2L 18778613 91 - 22036055 ----------------------GCCGAAA--CCAAGAGGCCGAGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUCUCGUCUCGUCUCUCCUCACCAACCGACUGGCAAACU---- ----------------------((((...--...((((((.((((.((..(((....((((........))))...))).))))))))))))..((.......)).)))).....---- ( -24.90, z-score = -1.28, R) >consensus ______________________GCCGAAA__CCAAGAGGCCGAGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUCUCGUCUCGCCUCGCCUCACCAACCGACUGGCAAACU____ ......................((((......)..(((((.(((.(((..(((....((((........))))...))).)))))))))))................)))......... (-14.64 = -16.42 + 1.78)

| Location | 19,084,758 – 19,084,848 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Shannon entropy | 0.37932 |
| G+C content | 0.53058 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -24.73 |
| Energy contribution | -24.27 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19084758 90 + 23011544 GACUUAUGGCUAUGAGCUGCCAAGUGGUCGGCGCUCU----------------CGGCCUCUU-GGUUUC--------GGUCAUUUUCCAAAGGCCAAAAUAAUGUGCGUAGCCAA--- ......((((((((.((.((((((.((((((.....)----------------))))).)))-)))...--------((((..........))))........)).)))))))).--- ( -34.70, z-score = -3.19, R) >droAna3.scaffold_12916 10838465 93 - 16180835 GACGUGUGCUUAUGAGCUGCCAAGUGGUCGGCGCUCA----------------UGGUCUCUUUGGUUUC--------GGCCAUUUUCCAAAAGCCAAAAUG-UGCGUACUGCCACUCA ((.(((.((.(((((((.(((........))))))))----------------))(((..((((((((.--------((.......))..))))))))..)-.)).....))))))). ( -26.90, z-score = -0.44, R) >droEre2.scaffold_4845 17402536 109 - 22589142 GACUUAUGGCUAUGAGCUGCCAAGUGGUCGCCGCUCUCAGGCUCUCGGCCUCUCGGCCUUUU-GGUCUC--------GGCCUUUUUCGAAUGGCCAAAAUAAUGUGCGUGGCCAAAGG ..(((.((((((((.((.(((.((.(((((..(((....)))...))))).)).))).((((-((((((--------((......))))..))))))))....)).))))))))))). ( -41.00, z-score = -1.95, R) >droYak2.chr2R 5600576 117 + 21139217 GACUUAUGGCUAUGAGCUGCCAAGUGGUCGCUGCUCUUCGCCUCUCGACCUCUCGGUCUCUA-GGUCUCUUGGUUUGGGCCUUUUUCGAAUGGCCAAAAUAAUGUGCGUAGCCAAAGG ..(((.((((((((.((.((((((.(..(((........)).....((((....))))....-)..).))))))...((((..........))))........)).))))))))))). ( -40.00, z-score = -2.31, R) >droSec1.super_7 2714537 93 + 3727775 GACUUAUGGCUAUGAGCUGCCAAGUGGUCGGCGCUCU----------------CGGCCUCUU-GGUUUC--------GGCCAUUUUCCAAAGGCCAAAAUAAUGUGCGUAGCCAACGG ......((((((((.((.((((((.((((((.....)----------------))))).)))-)))...--------((((..........))))........)).)))))))).... ( -37.40, z-score = -3.10, R) >droSim1.chr2L 18778649 93 + 22036055 GACUUAUGGCUAUGAGCUGCCAAGUGGUCGGCGCUCU----------------CGGCCUCUU-GGUUUC--------GGCCAUUUUCCAAAGGCCAAAAUAAUGUGCGUAGCCAACGG ......((((((((.((.((((((.((((((.....)----------------))))).)))-)))...--------((((..........))))........)).)))))))).... ( -37.40, z-score = -3.10, R) >consensus GACUUAUGGCUAUGAGCUGCCAAGUGGUCGGCGCUCU________________CGGCCUCUU_GGUUUC________GGCCAUUUUCCAAAGGCCAAAAUAAUGUGCGUAGCCAAAGG ......((((((((.((.((((((.(((((.......................))))).))).)))...........((((..........))))........)).)))))))).... (-24.73 = -24.27 + -0.47)
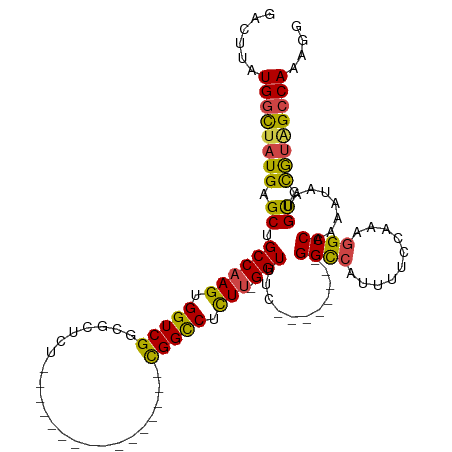
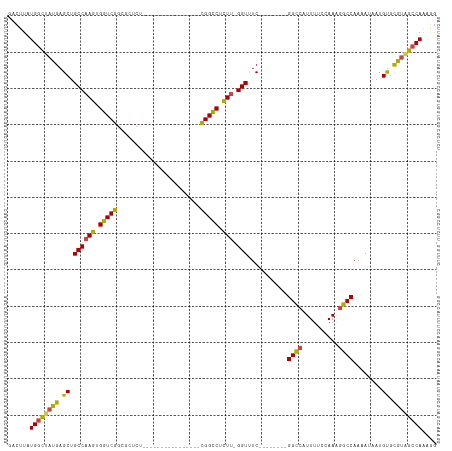
| Location | 19,084,758 – 19,084,848 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Shannon entropy | 0.37932 |
| G+C content | 0.53058 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -19.80 |
| Energy contribution | -21.21 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
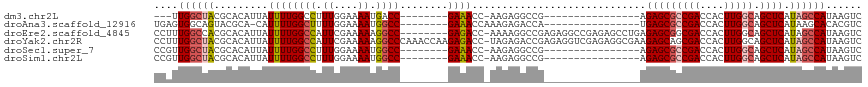
>dm3.chr2L 19084758 90 - 23011544 ---UUGGCUACGCACAUUAUUUUGGCCUUUGGAAAAUGACC--------GAAACC-AAGAGGCCG----------------AGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUC ---.((((((...........(((((((((((.........--------....))-.))))))))----------------)(((((((((....))))).)))).))))))...... ( -33.72, z-score = -4.27, R) >droAna3.scaffold_12916 10838465 93 + 16180835 UGAGUGGCAGUACGCA-CAUUUUGGCUUUUGGAAAAUGGCC--------GAAACCAAAGAGACCA----------------UGAGCGCCGACCACUUGGCAGCUCAUAAGCACACGUC ...(((((........-..((((((((.((....)).))))--------))))...........(----------------((((((((((....))))).))))))..)).)))... ( -30.70, z-score = -2.44, R) >droEre2.scaffold_4845 17402536 109 + 22589142 CCUUUGGCCACGCACAUUAUUUUGGCCAUUCGAAAAAGGCC--------GAGACC-AAAAGGCCGAGAGGCCGAGAGCCUGAGAGCGGCGACCACUUGGCAGCUCAUAGCCAUAAGUC .(((((((((.((......((((((((..........))))--------))))..-....((((....))))....)).)).((((.((.(.....).)).))))...)))).))).. ( -38.10, z-score = -2.35, R) >droYak2.chr2R 5600576 117 - 21139217 CCUUUGGCUACGCACAUUAUUUUGGCCAUUCGAAAAAGGCCCAAACCAAGAGACC-UAGAGACCGAGAGGUCGAGAGGCGAAGAGCAGCGACCACUUGGCAGCUCAUAGCCAUAAGUC .(((((((((.........(((.((((..........)))).)))..........-..(((.(((((.(((((....((.....))..))))).)))))...))).)))))).))).. ( -36.90, z-score = -2.85, R) >droSec1.super_7 2714537 93 - 3727775 CCGUUGGCUACGCACAUUAUUUUGGCCUUUGGAAAAUGGCC--------GAAACC-AAGAGGCCG----------------AGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUC ....((((((.((...((.((((((((.((....)).))))--------))))..-))...))..----------------.(((((((((....))))).)))).))))))...... ( -33.90, z-score = -2.84, R) >droSim1.chr2L 18778649 93 - 22036055 CCGUUGGCUACGCACAUUAUUUUGGCCUUUGGAAAAUGGCC--------GAAACC-AAGAGGCCG----------------AGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUC ....((((((.((...((.((((((((.((....)).))))--------))))..-))...))..----------------.(((((((((....))))).)))).))))))...... ( -33.90, z-score = -2.84, R) >consensus CCGUUGGCUACGCACAUUAUUUUGGCCUUUGGAAAAUGGCC________GAAACC_AAGAGGCCG________________AGAGCGCCGACCACUUGGCAGCUCAUAGCCAUAAGUC ....((((((.............(((((((...........................)))))))..................(((((((((....))))).)))).))))))...... (-19.80 = -21.21 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:17 2011