| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,808,444 – 1,808,554 |
| Length | 110 |
| Max. P | 0.551811 |

| Location | 1,808,444 – 1,808,554 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.92 |
| Shannon entropy | 0.68458 |
| G+C content | 0.40495 |
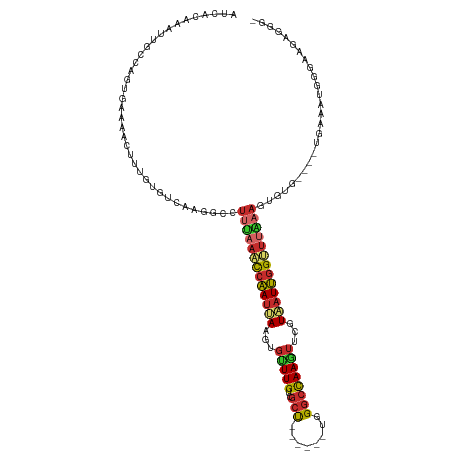
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -11.18 |
| Energy contribution | -11.44 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
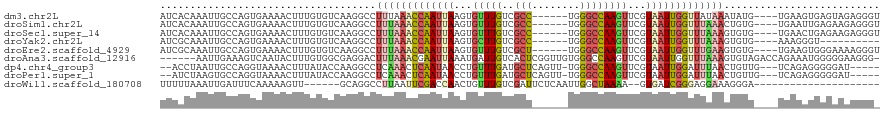
>dm3.chr2L 1808444 110 + 23011544 AUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCC------UGGGCCAAGUUCGUAAUUGGUUAUAAAUAUG----UGAAGUGAGUAGAGGGU .(((((.(((((((((...(((((.(.(((.((((...((((((......).)))))..)))------).)))))))))....))))))....))).))----))).............. ( -25.90, z-score = -0.95, R) >droSim1.chr2L 1791572 110 + 22036055 AUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCC------UGGGCCAAGUUCGUAAUUGGUUUAAACUGUG----UGAAUUGAGAAGAGGGU .((.(((.(..(((((.....(((((...)))))...((((((((((((.(..((((..((.------...))))))..).)))))))))))))))).)----..).))).))....... ( -26.70, z-score = -0.74, R) >droSec1.super_14 359126 110 - 2068291 AUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCC------UGGGCCAAGUUCGUAAUUGGUUUAAAGUGUG----UGAACUGAGAAGAGGGU .((((.........))))...((((..((..(.((((((((((((((((.(..((((..((.------...))))))..).)))))))))))))).)).----)..))...))))..... ( -29.10, z-score = -1.45, R) >droYak2.chr2L 1780470 100 + 22324452 AUCGCAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGCUUGUCGCC------UGGGCCAAGUUCGUAAUUGGUUUAAAGUGUG----AAAGGGU---------- .(((((....((((((....)))(((...))))))((((((((((((((...(((((..((.------...)))))))...))))))))))))))))))----)......---------- ( -32.60, z-score = -3.28, R) >droEre2.scaffold_4929 1849181 110 + 26641161 AUCGCAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCU------UGGGCCAAGUUCGUAAUUGGUUUGAAGUGUG----UGAAGUGGGAAAAGGGU .(((((....((((((....)))(((...))))))((((((((((((((.(..((((..((.------...))))))..).))))))))))))))..))----))).............. ( -27.60, z-score = -0.90, R) >droAna3.scaffold_12916 400772 113 - 16180835 ------AAUUGAAAGUCAAUACUUUGUGGCGAGGACUUUAAACGAAUUAAAUGAUUGUCACUCGGUUGUGGGCCAAGUUCGUAAUUGGUUUAAAGUGUAGACCAGAAAUGGGGGAAGGG- ------.((((.....)))).((((.(.....(.(((((((((.(((((.....(((((((......))))..))).....))))).))))))))).)...(((....)))).))))..- ( -26.20, z-score = -0.85, R) >dp4.chr4_group3 3188922 109 - 11692001 --ACCUAAUUGCCAGGUAAAACUUUAUACCAAGGCCUCAAACUCAAUAACCUGUUUGAUGCUCAGUU-UGGGCCAAGUUCGUAAUUGGAUUUAACUGUUG---UCAGAGGGGGAU----- --........(((.((((........))))..)))..............(((.((((((...(((((-....((((........))))....)))))..)---))))).)))...----- ( -27.60, z-score = -0.42, R) >droPer1.super_1 329984 109 - 10282868 --AUCUAAGUGCCAGGUAAAACUUUAUACCAAGGCCUCAAACUCAAUAACCUGUUUGAUGCUCAGUU-UGGGCCAAGUUCGUAAUUGGAUUUAACUGUUG---UCAGAGGGGGAU----- --.....((.(((.((((........))))..)))))............(((.((((((...(((((-....((((........))))....)))))..)---))))).)))...----- ( -28.30, z-score = -0.69, R) >droWil1.scaffold_180708 689711 91 - 12563649 UUUUUAAAUUGAUUUCAAAAAGUU------GCAGGCCUUAAUUCGACCAACUGUUUGUCGAUUCUCAAUUGGCUAAAA--GUGAUCGGGAGGAAAGGGA--------------------- ........(((....)))......------.....((((..(((..((.(((....((((((.....))))))....)--))....))..)))))))..--------------------- ( -14.60, z-score = 1.42, R) >consensus AUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCU______UGGGCCAAGUUCGUAAUUGGUUUAAAGUGUG____UGAAAUGGGAAGAGGG_ ....................................(((((((((((((...(((((..(((........))))))))...))))))))))))).......................... (-11.18 = -11.44 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:16 2011