| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,048,262 – 19,048,359 |
| Length | 97 |
| Max. P | 0.999927 |

| Location | 19,048,262 – 19,048,359 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.85 |
| Shannon entropy | 0.69429 |
| G+C content | 0.31197 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -12.41 |
| Energy contribution | -12.69 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.94 |
| SVM RNA-class probability | 0.999927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
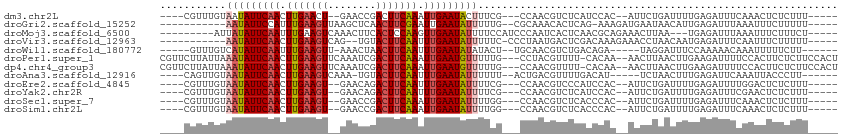
>dm3.chr2L 19048262 97 + 23011544 ----CGUUUGUAAUAUUCAACUUGAACU--GAACCGACUUCAAAUUGAAUACUUUCG---CCAACGUCUCAUCCAC--AUUCUGAUUUUGAGAUUUCAAACUCUCUUU----- ----.(((((...(((((((.(((((..--........))))).)))))))......---.....((((((.....--..........))))))..))))).......----- ( -15.46, z-score = -1.84, R) >droGri2.scaffold_15252 423429 94 + 17193109 -----------AAUAUUCCAUUUGAAGUUAAGCUCAACUUCGAAUUGAAUAUUUUUG--CGCAAACACUCAG-AAAGAUGAAUAACAUUGAGAUUUAAAUUUCUUUUU----- -----------(((((((.((((((((((......)))))))))).)))))))....--...........((-(((..(((((.........)))))..)))))....----- ( -17.50, z-score = -1.86, R) >droMoj3.scaffold_6500 23390397 96 - 32352404 ---------AUUAUAUUCAAUUUGAAGUCAAACUUCACUCCAAGUUGAAUAUUUUCCAUCCCAAUCACUCAACGCAGAAACUUAA---UGAGAUUUAAAUUUCUUUCU----- ---------...((((((((((((.(((........))).)))))))))))).......................(((((.((((---......)))).)))))....----- ( -15.60, z-score = -3.06, R) >droVir3.scaffold_12963 15129209 94 - 20206255 -----------AAUAUUCAACUUGAAGUCAG--UGUACUUCAAUUUGAAUAUUUUUC-CCCUAAUGACUCGACAAAGAAACCUAACAAUGAGAUUUCAAUUUCUUUUU----- -----------(((((((((.(((((((...--...))))))).)))))))))....-..................((((.((.......)).))))...........----- ( -15.60, z-score = -1.78, R) >droWil1.scaffold_180772 747294 94 - 8906247 -----GUUUGUCAUAUUCAAUUUGAAGUU-AAACUAACUUCAAUUUGAAUAUAUACU--UGCAACGUCUGACAGA-----UAGGAUUUCCAAAAACAAAUUUUUCUU------ -----(((((((((((((((.((((((((-(...))))))))).))))))))..((.--......))..))))))-----).((....)).................------ ( -22.20, z-score = -4.17, R) >droPer1.super_1 8168306 107 - 10282868 CGUUCUUAUUAAAUAUUCAACUUGAAGUUCAAAUCGACUUCAAAUUGAAUGUUUUUG---CCUACGUUUU-CACAA--AACUUAACUUGAAGAUUUUCCACUUCUCUUCCACU (((.......((((((((((.((((((((......)))))))).))))))))))...---...)))....-.....--..........(((((...........))))).... ( -20.72, z-score = -4.09, R) >dp4.chr4_group3 11053126 107 - 11692001 CGUUCUUAUUAAAUAUUCAACUUGAAGUUCAAAUCGACUUCAAAUUGAAUGUUUUUG---CCAACGUUUU-CACAA--AACUUAACUUGAAGAUUUUCCACUUCUCUUCCACU ((((......((((((((((.((((((((......)))))))).))))))))))...---..))))....-.....--..........(((((...........))))).... ( -21.10, z-score = -4.04, R) >droAna3.scaffold_12916 10802256 95 - 16180835 ----CAGUUGUAAUAUUCAACUUGAAGUCAAA-UGUACUUCAAUUUGAAUAUUUUUU--ACUGACGUUUUGACAU-----UCUAACUUUGAGAUUCAAAUUACCCUU------ ----((((...(((((((((.((((((((...-.).))))))).)))))))))....--))))....(((((..(-----((.......)))..)))))........------ ( -21.80, z-score = -3.69, R) >droEre2.scaffold_4845 17366809 97 - 22589142 ----CGUUUGUAAUAUUCAACUUGAAGU--GAACAGACUUCAAUUUGAAUAUUUUCG---CCAACGUCCCAUCCAC--AUUCUGAUUUUGAGAUUUUGGACUCUCUUU----- ----((((.(((((((((((.(((((((--......))))))).)))))))))...)---).)))).....((((.--..(((.......)))...))))........----- ( -22.90, z-score = -3.02, R) >droYak2.chr2R 5564103 97 + 21139217 ----CGUUUGUAAUAUUCAACUUGAAGU--GAACAGACUUCAAUUUGAAUAUUUUCG---CCAACGUCUCAUCCAC--AUUCUGAUUUUGAGAUUUCGAACUCUCUUU----- ----.(((((.(((((((((.(((((((--......))))))).)))))))))....---.....((((((.....--..........))))))..))))).......----- ( -22.56, z-score = -2.98, R) >droSec1.super_7 2678253 97 + 3727775 ----CGUUUGUAAUAUUCAACUUGAAGU--GAACCGACUUCAAAUUGAAUAUUUUGG---CCAACGUCUCACCCAC--AUUCUGAUUUUGAGAUUUCAAACUCUCUUU----- ----.(((((.(((((((((.(((((((--......))))))).)))))))))....---.....((((((.....--..........))))))..))))).......----- ( -21.96, z-score = -3.15, R) >droSim1.chr2L 18742946 97 + 22036055 ----CGUUUGUAAUAUUCAACUUGAAGU--GAACCGACUUCAAAUUGAAUAUUUUGG---CCAACGUCUCACCCAC--AUUCUGAUUUUGAGAUUUCAAACUCUCUUU----- ----.(((((.(((((((((.(((((((--......))))))).)))))))))....---.....((((((.....--..........))))))..))))).......----- ( -21.96, z-score = -3.15, R) >consensus ____CGUUUGUAAUAUUCAACUUGAAGU__AAACCGACUUCAAAUUGAAUAUUUUCG___CCAACGUCUCACCCAA__AUUCUAAUUUUGAGAUUUCAAACUCUCUUU_____ ...........(((((((((.(((((((........))))))).)))))))))............................................................ (-12.41 = -12.69 + 0.29)

| Location | 19,048,262 – 19,048,359 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 65.85 |
| Shannon entropy | 0.69429 |
| G+C content | 0.31197 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -13.24 |
| Energy contribution | -13.56 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
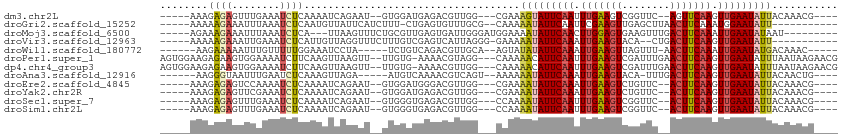
>dm3.chr2L 19048262 97 - 23011544 -----AAAGAGAGUUUGAAAUCUCAAAAUCAGAAU--GUGGAUGAGACGUUGG---CGAAAGUAUUCAAUUUGAAGUCGGUUC--AGUUCAAGUUGAAUAUUACAAACG---- -----.......(((((...(((((...(((....--.))).)))))((....---))..(((((((((((((((.(......--).))))))))))))))).))))).---- ( -23.20, z-score = -2.05, R) >droGri2.scaffold_15252 423429 94 - 17193109 -----AAAAAGAAAUUUAAAUCUCAAUGUUAUUCAUCUUU-CUGAGUGUUUGCG--CAAAAAUAUUCAAUUCGAAGUUGAGCUUAACUUCAAAUGGAAUAUU----------- -----........................(((((......-.(((((((((...--...)))))))))....(((((((....))))))).....)))))..----------- ( -16.40, z-score = -1.23, R) >droMoj3.scaffold_6500 23390397 96 + 32352404 -----AGAAAGAAAUUUAAAUCUCA---UUAAGUUUCUGCGUUGAGUGAUUGGGAUGGAAAAUAUUCAACUUGGAGUGAAGUUUGACUUCAAAUUGAAUAUAAU--------- -----....((((((((((......---))))))))))...(..(....)..)........((((((((.(((((((........))))))).))))))))...--------- ( -21.00, z-score = -1.93, R) >droVir3.scaffold_12963 15129209 94 + 20206255 -----AAAAAGAAAUUGAAAUCUCAUUGUUAGGUUUCUUUGUCGAGUCAUUAGGG-GAAAAAUAUUCAAAUUGAAGUACA--CUGACUUCAAGUUGAAUAUU----------- -----.....((....(((((((.......)))))))....))............-....(((((((((.(((((((...--...))))))).)))))))))----------- ( -21.00, z-score = -2.10, R) >droWil1.scaffold_180772 747294 94 + 8906247 ------AAGAAAAAUUUGUUUUUGGAAAUCCUA-----UCUGUCAGACGUUGCA--AGUAUAUAUUCAAAUUGAAGUUAGUUU-AACUUCAAAUUGAAUAUGACAAAC----- ------.......((((((.(((((........-----....)))))....)))--))).(((((((((.(((((((((...)-)))))))).)))))))))......----- ( -19.40, z-score = -2.22, R) >droPer1.super_1 8168306 107 + 10282868 AGUGGAAGAGAAGUGGAAAAUCUUCAAGUUAAGUU--UUGUG-AAAACGUAGG---CAAAAACAUUCAAUUUGAAGUCGAUUUGAACUUCAAGUUGAAUAUUUAAUAAGAACG ....(((((....(....).)))))..(((..(((--((((.-...))).)))---)..(((.((((((((((((((........)))))))))))))).)))......))). ( -24.40, z-score = -2.46, R) >dp4.chr4_group3 11053126 107 + 11692001 AGUGGAAGAGAAGUGGAAAAUCUUCAAGUUAAGUU--UUGUG-AAAACGUUGG---CAAAAACAUUCAAUUUGAAGUCGAUUUGAACUUCAAGUUGAAUAUUUAAUAAGAACG ....(((((....(....).))))).......(((--((.((-((...(((..---....)))((((((((((((((........)))))))))))))).))))...))))). ( -24.70, z-score = -2.28, R) >droAna3.scaffold_12916 10802256 95 + 16180835 ------AAGGGUAAUUUGAAUCUCAAAGUUAGA-----AUGUCAAAACGUCAGU--AAAAAAUAUUCAAAUUGAAGUACA-UUUGACUUCAAGUUGAAUAUUACAACUG---- ------........(((((...((.......))-----...)))))....((((--....(((((((((.(((((((.(.-...)))))))).)))))))))...))))---- ( -19.40, z-score = -2.43, R) >droEre2.scaffold_4845 17366809 97 + 22589142 -----AAAGAGAGUCCAAAAUCUCAAAAUCAGAAU--GUGGAUGGGACGUUGG---CGAAAAUAUUCAAAUUGAAGUCUGUUC--ACUUCAAGUUGAAUAUUACAAACG---- -----.......(.((((..(((((...(((....--.))).)))))..))))---)...(((((((((.(((((((......--))))))).))))))))).......---- ( -23.50, z-score = -2.35, R) >droYak2.chr2R 5564103 97 - 21139217 -----AAAGAGAGUUCGAAAUCUCAAAAUCAGAAU--GUGGAUGAGACGUUGG---CGAAAAUAUUCAAAUUGAAGUCUGUUC--ACUUCAAGUUGAAUAUUACAAACG---- -----...((((........))))..........(--((.((((...)))).)---))..(((((((((.(((((((......--))))))).))))))))).......---- ( -22.40, z-score = -1.68, R) >droSec1.super_7 2678253 97 - 3727775 -----AAAGAGAGUUUGAAAUCUCAAAAUCAGAAU--GUGGGUGAGACGUUGG---CCAAAAUAUUCAAUUUGAAGUCGGUUC--ACUUCAAGUUGAAUAUUACAAACG---- -----.......(((((...(((((...(((....--.))).)))))......---....(((((((((((((((((......--))))))))))))))))).))))).---- ( -29.10, z-score = -4.05, R) >droSim1.chr2L 18742946 97 - 22036055 -----AAAGAGAGUUUGAAAUCUCAAAAUCAGAAU--GUGGGUGAGACGUUGG---CCAAAAUAUUCAAUUUGAAGUCGGUUC--ACUUCAAGUUGAAUAUUACAAACG---- -----.......(((((...(((((...(((....--.))).)))))......---....(((((((((((((((((......--))))))))))))))))).))))).---- ( -29.10, z-score = -4.05, R) >consensus _____AAAGAGAGUUUGAAAUCUCAAAAUCAGAAU__GUGGGUGAGACGUUGG___CAAAAAUAUUCAAUUUGAAGUCGAUUC__ACUUCAAGUUGAAUAUUACAAACG____ ........((((........))))....................................(((((((((.(((((((........))))))).)))))))))........... (-13.24 = -13.56 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:13 2011