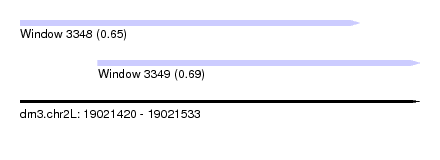
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,021,420 – 19,021,533 |
| Length | 113 |
| Max. P | 0.693489 |

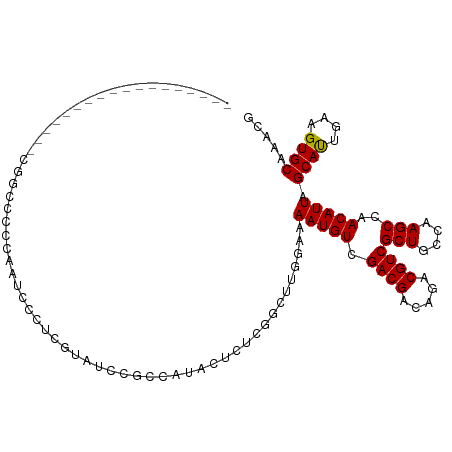
| Location | 19,021,420 – 19,021,516 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.41 |
| Shannon entropy | 0.42075 |
| G+C content | 0.54718 |
| Mean single sequence MFE | -24.29 |
| Consensus MFE | -13.41 |
| Energy contribution | -13.22 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19021420 96 + 23011544 ------------------CGACCCCCAAUCCCUUGUAUCCGCUAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACG ------------------..............((((((..((((.......(((((((.....(.(((((.....))))))..))))))).....))))......))))))... ( -24.10, z-score = -1.95, R) >droSim1.chr2L 18714909 96 + 22036055 ------------------CGGCCCCCAAUCCCUCGUAUCCGCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACG ------------------.(((..................)))........(((((((.....(.(((((.....))))))..))))))).......((((....))))..... ( -25.97, z-score = -1.72, R) >droSec1.super_7 2651966 96 + 3727775 ------------------CGGCCCCCAAUCCCUCGUAUCCGCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACG ------------------.(((..................)))........(((((((.....(.(((((.....))))))..))))))).......((((....))))..... ( -25.97, z-score = -1.72, R) >droYak2.chr2R 5538682 92 + 21139217 ----------------------CACCAAGCCCCCGUAUCCGCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCACUGAAGUGCAAACG ----------------------..(((((((...((((.....))))....)))))))..(((((.((((.....))))(((....)))..))))).((((....))))..... ( -27.80, z-score = -3.05, R) >droEre2.scaffold_4845 17341536 96 - 22589142 ------------------CAGCCCCCCAUCCCGCGCAUACGCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCACUGAAGUGCAAACG ------------------..............(((....))).........(((((((.....(.(((((.....))))))..))))))).......((((....))))..... ( -27.80, z-score = -2.41, R) >droAna3.scaffold_12916 10776689 79 - 16180835 ------------------------CCAAAAUUUCGAAUCCUGCAU-----------GGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUCGAAGUGCAAACG ------------------------.....(((((((.(((.....-----------))).(((((.((((.....))))(((....)))..)))))....)))))))....... ( -18.30, z-score = -1.27, R) >droPer1.super_1 8141164 111 - 10282868 CAGUCCCACGCCCUUACCCUUGCUGCAACCCGGCCUUUCCCCCAUCCACACGG---AGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACG .........((.(((.....(((((......(((.(((((...........))---)))....(.(((((.....))))))......))).....)))))...))).))..... ( -20.90, z-score = -0.00, R) >dp4.chr4_group3 11026300 111 - 11692001 CAGUCCCACGCCCUUACCCUUGCUGCAACCCGGCCUCUCCCCCAUCCACACGG---AGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACG .........((.(((.....(((((......(((.(((((...........))---)))....(.(((((.....))))))......))).....)))))...))).))..... ( -23.50, z-score = -0.84, R) >consensus __________________CGGCCCCCAAUCCCUCGUAUCCGCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACG ............................................................(((((.((((.....))))(((....)))..))))).((((....))))..... (-13.41 = -13.22 + -0.19)

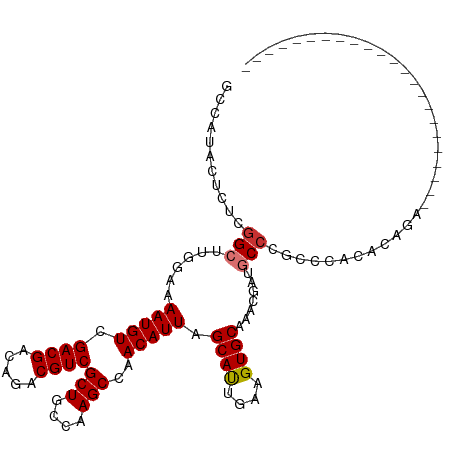
| Location | 19,021,442 – 19,021,533 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Shannon entropy | 0.37054 |
| G+C content | 0.54586 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -16.06 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19021442 91 + 23011544 GCUAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACGAUGCCCGCCCACACAGA----------------------- ((.........(((((((.....(.(((((.....))))))..))))))).......((((((.........))))))..)).........----------------------- ( -25.80, z-score = -1.77, R) >droSim1.chr2L 18714931 91 + 22036055 GCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACGAUGCCCGCCCACACAGA----------------------- ((.........(((((((.....(.(((((.....))))))..))))))).......((((((.........))))))..)).........----------------------- ( -25.40, z-score = -1.63, R) >droSec1.super_7 2651988 91 + 3727775 GCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACGAUGCCCGCCCACACAGA----------------------- ((.........(((((((.....(.(((((.....))))))..))))))).......((((((.........))))))..)).........----------------------- ( -25.40, z-score = -1.63, R) >droYak2.chr2R 5538700 91 + 21139217 GCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCACUGAAGUGCAAACGAUGCCCGCCCACACAGA----------------------- ((.........(((((((.....(.(((((.....))))))..))))))).......((((....))))...........)).........----------------------- ( -26.10, z-score = -1.82, R) >droEre2.scaffold_4845 17341558 91 - 22589142 GCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCACUGAAGUGCAAACGAUGCCCGCCCACACAGA----------------------- ((.........(((((((.....(.(((((.....))))))..))))))).......((((....))))...........)).........----------------------- ( -26.10, z-score = -1.82, R) >droAna3.scaffold_12916 10776704 96 - 16180835 ----------CUGCAUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUCGAAGUGCAAACGACGCCCACCCAUACAGAUACUUACCAACACAC-------- ----------(((.((((....(((((..(((....)))(((....)))........((((....))))...))))).....)))).)))................-------- ( -20.40, z-score = -1.74, R) >droPer1.super_1 8141204 111 - 10282868 CCCAUCCACACGG---AGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACGACGCCCACACACACACAGACAUUUGCCCGCACCCCGCCGC ..........(((---.(.(((((((((((.....))))(((....)))........((((....))))......................))))))).))))........... ( -22.20, z-score = -0.87, R) >dp4.chr4_group3 11026340 111 - 11692001 CCCAUCCACACGG---AGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACGACGCCCACACACACACAGACAUUUGCCCGCACCCCGCCGC ..........(((---.(.(((((((((((.....))))(((....)))........((((....))))......................))))))).))))........... ( -22.20, z-score = -0.87, R) >consensus GCCAUACUCUCGGCUUGGAAAAUGUCGACGACAGACGUCGCUGCCAAGCCAACAUUAGCAUUGAAGUGCAAACGAUGCCCGCCCACACAGA_______________________ ...........(((.(((.....(.(((((.....))))))..))).))).......((((....))))............................................. (-16.06 = -16.75 + 0.69)

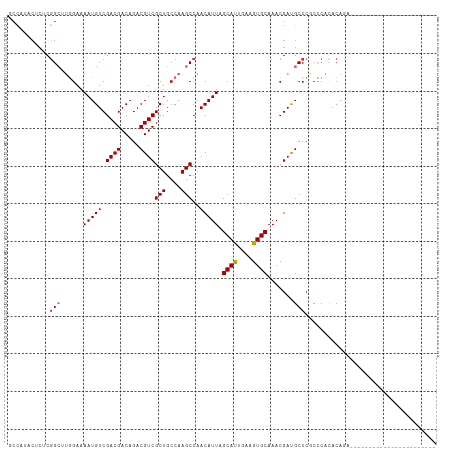
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:08 2011