| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,800,504 – 1,800,557 |
| Length | 53 |
| Max. P | 0.958269 |

| Location | 1,800,504 – 1,800,557 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 64.65 |
| Shannon entropy | 0.62706 |
| G+C content | 0.36429 |
| Mean single sequence MFE | -10.76 |
| Consensus MFE | -4.92 |
| Energy contribution | -5.44 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
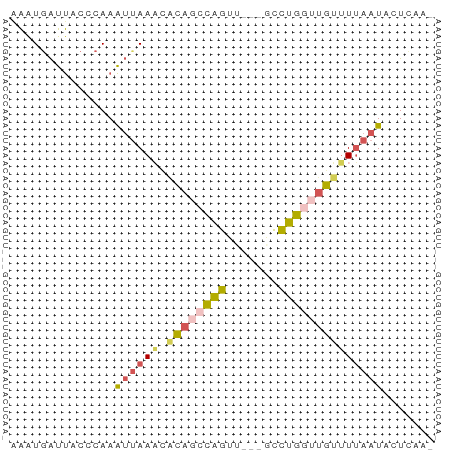
>dm3.chr2L 1800504 53 - 23011544 AAAUGAUUACCCAAAUGAACCACAGCCAGUU---GCCUGGUUGUUUUAAUACUCAA- ...............((((..((((((((..---..))))))))))))........- ( -10.30, z-score = -1.73, R) >droSim1.chr2L 1789116 53 - 22036055 UAAUGAUUACCCAAAUUAAAUACAGCCAGUU---GCCUGGUUGUUUUAAUACUCAA- ..............((((((.((((((((..---..))))))))))))))......- ( -13.00, z-score = -3.45, R) >droSec1.super_14 1752150 53 - 2068291 UAAUGAUUACGCAAAUUAAAUACACCCAGUU---CCCUGGUUGUUUUAAUACUCAA- ..............((((((.(((.((((..---..)))).)))))))))......- ( -8.80, z-score = -2.16, R) >droEre2.scaffold_4929 1846755 53 - 26641161 AAAUGGUUAC-CAAAAUAAAGGUAGCCAGUU---UCCUGCUUGUUUUAAUACUUCGA ...(((((((-(........))))))))...---....................... ( -11.80, z-score = -2.17, R) >droVir3.scaffold_12734 107219 54 - 510240 GGGUAACCACCCAAGUUAAUCACGAGUUACCAAGGUUGGGGGGGGUAAAUAAUA--- .....(((.(((...((((((............)))))).))))))........--- ( -9.90, z-score = 1.18, R) >consensus AAAUGAUUACCCAAAUUAAACACAGCCAGUU___GCCUGGUUGUUUUAAUACUCAA_ ..............((((((.((((((((.......))))))))))))))....... ( -4.92 = -5.44 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:15 2011