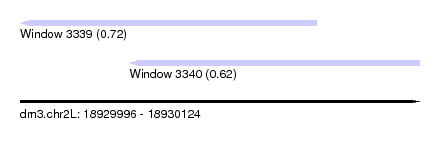
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,929,996 – 18,930,124 |
| Length | 128 |
| Max. P | 0.719507 |

| Location | 18,929,996 – 18,930,091 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.74 |
| Shannon entropy | 0.15695 |
| G+C content | 0.44877 |
| Mean single sequence MFE | -13.68 |
| Consensus MFE | -11.32 |
| Energy contribution | -11.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
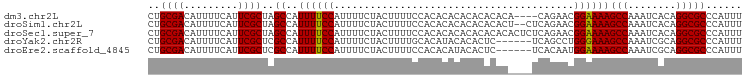
>dm3.chr2L 18929996 95 - 23011544 CUGCGACAUUUUCAUUCGCUAGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACACA----CAGAACGGAAAAGCCAAAUCACAGGCGCCCAUUU ..((((.........))))..((..((((((...((((.......................----.)))).))))))(((........)))))...... ( -12.46, z-score = -2.19, R) >droSim1.chr2L 18621845 97 - 22036055 CUGCGACAUUUUCAUUCGCUAGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACACU--CUCAGAACGGAAAAGCCAAAUCACAGGCGCCCAUUU ..((((.........))))..((..((((((...((((.......................--...)))).))))))(((........)))))...... ( -12.37, z-score = -1.83, R) >droSec1.super_7 2558585 99 - 3727775 CUGCGACAUUUUCAUUCGCUAGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACACACUCUCAGAACGGAAAAGCCAAAUCACAGGCGCCCAUUU ..((((.........))))..((..((((((...((((............................)))).))))))(((........)))))...... ( -12.29, z-score = -1.89, R) >droYak2.chr2R 5446780 93 - 21139217 CUGCGACAUUUUCAUUCGCUCGCCAUUUUCCAUUUUCUACUUUUGCACAUACACACUC------UCAGCCUGGGAAAGCCAAAUCGCAGGCGCCCAUUU ..((((.........)))).......................................------...(((((.((........)).)))))........ ( -14.80, z-score = -0.10, R) >droEre2.scaffold_4845 17242002 93 + 22589142 CUGCGACAUUUUCAUUCGCUCGCCAUUUUCCAUUUUCUACUUUUCCACACAUACACUC------UCACAAUGGAAAAGCCAAAUCGCAGGCGCCCAUUU ..((((.........)))).((((.(((((((((........................------....)))))))))((......)).))))....... ( -16.49, z-score = -2.88, R) >consensus CUGCGACAUUUUCAUUCGCUAGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACAC____UCAGAACGGAAAAGCCAAAUCACAGGCGCCCAUUU ..((((.........))))..((..((((((........................................))))))(((........)))))...... (-11.32 = -11.16 + -0.16)
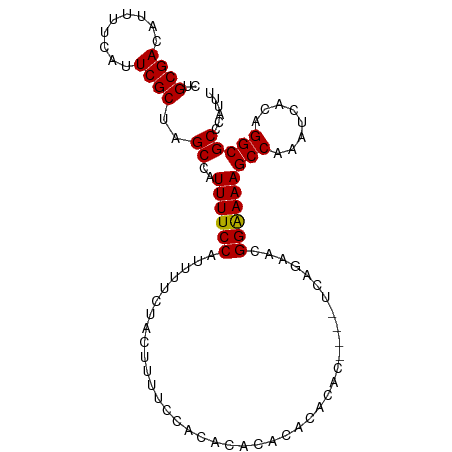
| Location | 18,930,031 – 18,930,124 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.28 |
| Shannon entropy | 0.21305 |
| G+C content | 0.40882 |
| Mean single sequence MFE | -10.59 |
| Consensus MFE | -9.31 |
| Energy contribution | -8.90 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
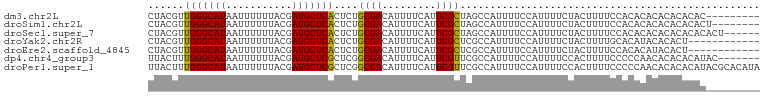
>dm3.chr2L 18930031 93 - 23011544 CUACGUUGGGCAUAAUUUUUUACGAUGCUCACUCUGCGACAUUUUCAUUCGCUAGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACAC--------- ......(((((((...........)))))))....((((.........)))).........................................--------- ( -9.70, z-score = -1.31, R) >droSim1.chr2L 18621881 94 - 22036055 CUACGUUGGGCAUAAUUUUUUACGAUGCUCACUCUGCGACAUUUUCAUUCGCUAGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACACU-------- ......(((((((...........)))))))....((((.........))))..........................................-------- ( -9.70, z-score = -1.23, R) >droSec1.super_7 2558621 96 - 3727775 CUACGUUGGGCAUAAUUUUUUACGAUGCUCACUCUGCGACAUUUUCAUUCGCUAGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACACACU------ ......(((((((...........)))))))....((((.........))))............................................------ ( -9.70, z-score = -1.33, R) >droYak2.chr2R 5446816 90 - 21139217 CUACGUUGGGCAUAAUUUUUUACGAUGCUCACUCUGCGACAUUUUCAUUCGCUCGCCAUUUUCCAUUUUCUACUUUUGCACAUACACACU------------ ......(((((((...........)))))))....((((.........))))......................................------------ ( -9.70, z-score = -0.66, R) >droEre2.scaffold_4845 17242038 90 + 22589142 CUACGUUGGGCAUAAUUUUUUACGAUGCUCACUCUGCGACAUUUUCAUUCGCUCGCCAUUUUCCAUUUUCUACUUUUCCACACAUACACU------------ ......(((((((...........)))))))....((((.........))))......................................------------ ( -9.70, z-score = -1.29, R) >dp4.chr4_group3 2295328 95 - 11692001 UUACUUUGGGCAUAAUUUUUUACGAUGCUCGCUCGGCGACAUUUUCAUUCGUUCGCCAUUUUCCAUUUUCCACUUUUCCCCCAACACACACAUAC------- .......((((((...........))))))....(((((.............)))))......................................------- ( -12.82, z-score = -1.62, R) >droPer1.super_1 3767584 102 - 10282868 UUACUUUGGGCAUAAUUUUUUACGAUGCUCGCUCGGCGACAUUUUCAUUCGUUCGCCAUUUUCCAUUUUCCACUUUUCCCCCAACACACACAUACGCACAUA .......((((((...........))))))....(((((.............)))))............................................. ( -12.82, z-score = -1.04, R) >consensus CUACGUUGGGCAUAAUUUUUUACGAUGCUCACUCUGCGACAUUUUCAUUCGCUCGCCAUUUUCCAUUUUCUACUUUUCCACACACACACACAC_________ ......(((((((...........)))))))....((((.........)))).................................................. ( -9.31 = -8.90 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:51:01 2011