
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,929,577 – 18,929,683 |
| Length | 106 |
| Max. P | 0.653118 |

| Location | 18,929,577 – 18,929,683 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Shannon entropy | 0.42677 |
| G+C content | 0.45874 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.653118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
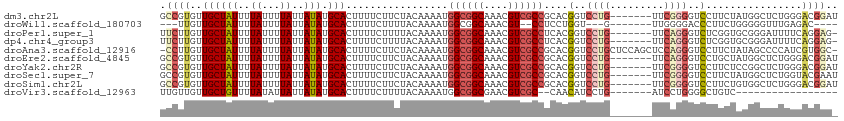
>dm3.chr2L 18929577 106 - 23011544 GCCGUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUG-------UUCGGGGUCCUUCUAUGGCUCUGGGACGGAU (((((((((((((..((...))..))).))).................((((((....))))))))))))).(((-------(((.((..((......))..)).)))))).. ( -34.70, z-score = -2.94, R) >droWil1.scaffold_180703 2289490 94 + 3946847 ---UUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGU--CCUCCUGGU---G-------UUGGGGACCCUUCUGGGGGUUUGAGAC---- ---(((((((((((((...........................))))))))))))).((.--.((((..(.---.-------..((....))..)..))))..))....---- ( -24.03, z-score = -1.26, R) >droPer1.super_1 3766810 105 - 10282868 UUCUUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCGCCUCACGGUCCUG-------UUCAGGGUCUCGGUGCGGGAUUUUCAGGAG- .(((((...((....(((......)))((((((...............((((((....))))))....(..(((.-------...)))..)..)))))))).....))))).- ( -26.30, z-score = -1.81, R) >dp4.chr4_group3 2294542 105 - 11692001 UUCUUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCAAACGUCGCCUCACGGUCCUG-------UUCAGGGUCUCGGUGCGGGAUUUUCAGGAG- .(((((...((....(((......)))((((((...............((((((....))))))....(..(((.-------...)))..)..)))))))).....))))).- ( -26.30, z-score = -1.81, R) >droAna3.scaffold_12916 1937938 111 - 16180835 -CCUUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUGCUCCAGCUCCAGGGUCCUUCUAUAGCCCCAUCGUGGC- -..(((((((((((((...........................))))))))))))).....(((((..((..(((...)))..)).((((.........))))....)))))- ( -26.63, z-score = -0.99, R) >droEre2.scaffold_4845 17241563 106 + 22589142 GCCGUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUG-------UUCAGGGUCCUGCUAUGGCUCUGGGACGGAU (((((((((((((..((...))..))).))).................((((((....))))))))))))).(((-------(((((((.((......)))))).)))))).. ( -34.70, z-score = -2.62, R) >droYak2.chr2R 5446349 106 - 21139217 GCCGUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUG-------UUCGGGGUCCUUCUCCGGCUCUGGGACGGAU (((((((((((((..((...))..))).))).................((((((....))))))))))))).(((-------(((.((..((......))..)).)))))).. ( -34.10, z-score = -2.43, R) >droSec1.super_7 2558142 106 - 3727775 GCCGUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUG-------UUCGGGGUCCUUCUAUGGCUCUGGUACGAAU (((((((((((((..((...))..))).))).................((((((....)))))))))))))..((-------(((((((.((......)))))))).)))... ( -30.90, z-score = -2.39, R) >droSim1.chr2L 18621400 106 - 22036055 GCCGUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUG-------UUCGGGGUCCUUCUGUGGCUCUGGGACGGAU (((((((((((((..((...))..))).))).................((((((....))))))))))))).(((-------(((.((..((......))..)).)))))).. ( -34.70, z-score = -2.62, R) >droVir3.scaffold_12963 9560965 87 - 20206255 UUGUUGUUGCUGUUUUAUAUUAUUAUAUGCACUUUUCUUUUACAAAAUGGCGGCGAACGUCGC--CAACAUCCUG-------AUCCUGGGGCUGUC----------------- .......(((.....((((....)))).)))................(((((((....)))))--))(((((((.-------.....)))).))).----------------- ( -17.80, z-score = -0.40, R) >consensus GCCGUGUUGCUAUUUUAUUUUAUUAUAUGCACUUUUCUUCUACAAAAUGGCGGCAAACGUCGCCGCACGGUCCUG_______UUCGGGGUCCUUCUAUGGCUCUGGGACGGA_ .((((..((((((..((...))..))).))).................((((((....))))))....(..((((.........))))..)................)))).. (-13.72 = -14.46 + 0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:59 2011