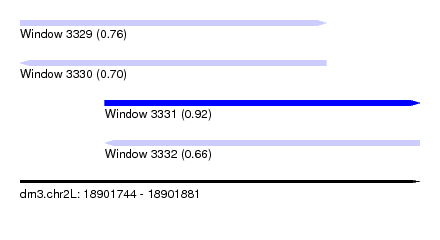
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,901,744 – 18,901,881 |
| Length | 137 |
| Max. P | 0.924770 |

| Location | 18,901,744 – 18,901,849 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.10 |
| Shannon entropy | 0.23116 |
| G+C content | 0.50816 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -24.95 |
| Energy contribution | -26.38 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
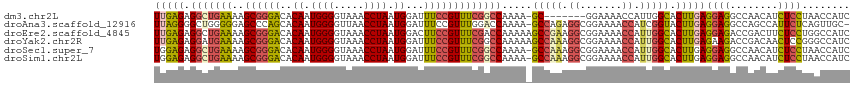
>dm3.chr2L 18901744 105 + 23011544 UCUUCUCCCGCAAUUGCAAACUGCAGCGCUUGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA--------GCGGAAAACCAUUG .......((((..((((.....)))).........(((((((..((((((..((.((((.....)))).))...)))))))))))))....--------)))).......... ( -34.70, z-score = -2.04, R) >droAna3.scaffold_12916 1914066 112 + 16180835 UCUUGGCCCACAAAUGCAAACUGCAGCGGUUAGGGGCUGGGGGAGCCCAGCACAAUGGGGUUAACCUAAUGGAUUUCCGUUUGGACCAAAA-GCCAGAGGCGGAAAACCAUCG ((((((((......(((.....)))..))))))))((((((....))))))....((((.....))))((((.(((((((((((.......-.))))..))))))).)))).. ( -42.20, z-score = -1.72, R) >droEre2.scaffold_4845 17213073 113 - 22589142 UCUUCUCCCUCAACUGCAAACUGCAUCGCUUGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGACUUCCGUUUCGACCAAAAAGCCGAAGGCGGAAAACCAUUG .....((((((((.(((.....)))....))))...(((....)))))))..((((((......((....))..((((((((((.(......).)))).))))))..)))))) ( -31.40, z-score = -1.23, R) >droYak2.chr2R 5418069 113 + 21139217 UCUUCUCCCUCAACUGCAAACUGCAGCGCUUGAGAGGAUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAAAGCCAAAGGCGGAAAACCAUUG ..(((((((((((((((.....))))...))))).))).)))..............(((.....)))(((((.(((((((((.(((......)))..))))))))).))))). ( -36.80, z-score = -2.40, R) >droSec1.super_7 2530538 112 + 3727775 UCUUCUCCCGCAACUGCAAACUGCAGCGCUGGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA-GCCAAAGGCGGAAAACCAUUG ...(((((.((..((((.....)))).)).)))))(((((((..((((((..((.((((.....)))).))...)))))))))))))....-(((...)))((....)).... ( -43.20, z-score = -3.41, R) >droSim1.chr2L 18591989 112 + 22036055 UCUUCUCCCGCAACUGCAAACUGCAGCACUGGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA-GCCAAAGGCGGAAAACCAUUG ...(((((.(...((((.....))))..).)))))(((((((..((((((..((.((((.....)))).))...)))))))))))))....-(((...)))((....)).... ( -39.80, z-score = -2.84, R) >consensus UCUUCUCCCGCAACUGCAAACUGCAGCGCUUGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA_GCCAAAGGCGGAAAACCAUUG ...((((......((((.....)))).....))))(((((((..((((((..((.((((.....)))).))...))))))))))))).....(((...)))((....)).... (-24.95 = -26.38 + 1.43)

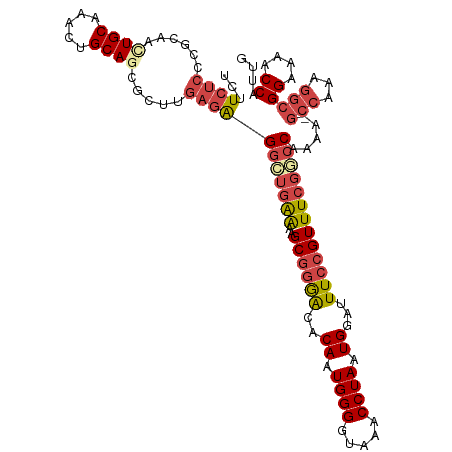
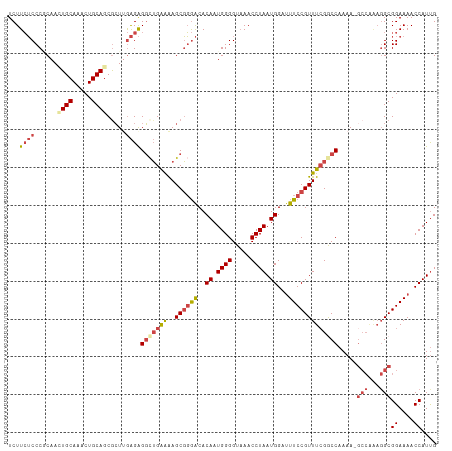
| Location | 18,901,744 – 18,901,849 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Shannon entropy | 0.23116 |
| G+C content | 0.50816 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -24.49 |
| Energy contribution | -25.52 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18901744 105 - 23011544 CAAUGGUUUUCCGC--------UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCAAGCGCUGCAGUUUGCAAUUGCGGGAGAAGA .(((((.((((((.--------((((....)))))))))).))))).((..(((......)))..)).((((((.((.......)).(((((((.....))))))))))))). ( -31.20, z-score = -0.91, R) >droAna3.scaffold_12916 1914066 112 - 16180835 CGAUGGUUUUCCGCCUCUGGC-UUUUGGUCCAAACGGAAAUCCAUUAGGUUAACCCCAUUGUGCUGGGCUCCCCCAGCCCCUAACCGCUGCAGUUUGCAUUUGUGGGCCAAGA .(((((.((((((....((((-.....).)))..)))))).))))).(((............((((((....))))))......(((((((.....)))...))))))).... ( -34.90, z-score = -0.31, R) >droEre2.scaffold_4845 17213073 113 + 22589142 CAAUGGUUUUCCGCCUUCGGCUUUUUGGUCGAAACGGAAGUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCAAGCGAUGCAGUUUGCAGUUGAGGGAGAAGA .(((((.((((((..(((((((....))))))).)))))).))))).((..(((......)))..)).((((((..((((....((((......))))....)))))))))). ( -38.70, z-score = -2.47, R) >droYak2.chr2R 5418069 113 - 21139217 CAAUGGUUUUCCGCCUUUGGCUUUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAUCCUCUCAAGCGCUGCAGUUUGCAGUUGAGGGAGAAGA .(((((.((((((..(((((((....))))))).)))))).))))).((..(((......)))..))..(((((.(((.((((...(((((.....))))))))))))))))) ( -42.90, z-score = -3.82, R) >droSec1.super_7 2530538 112 - 3727775 CAAUGGUUUUCCGCCUUUGGC-UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCCAGCGCUGCAGUUUGCAGUUGCGGGAGAAGA .(((((.((((((..((((((-.....)))))).)))))).))))).((..(((......)))..))(((....))).(((((.(((((((.....)))).))).)))))... ( -43.40, z-score = -3.38, R) >droSim1.chr2L 18591989 112 - 22036055 CAAUGGUUUUCCGCCUUUGGC-UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCCAGUGCUGCAGUUUGCAGUUGCGGGAGAAGA .(((((.((((((..((((((-.....)))))).)))))).))))).((..(((......)))..))(((....))).(((((...(((((.....)))))....)))))... ( -41.20, z-score = -2.95, R) >consensus CAAUGGUUUUCCGCCUUUGGC_UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCAAGCGCUGCAGUUUGCAGUUGCGGGAGAAGA .(((((.((((((..((((((......)))))).)))))).))))).......((((((((((..(.((....................)).)..))))).)).)))...... (-24.49 = -25.52 + 1.03)
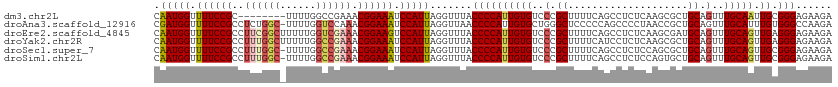
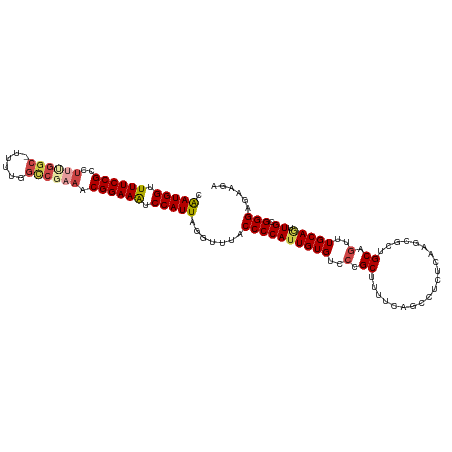
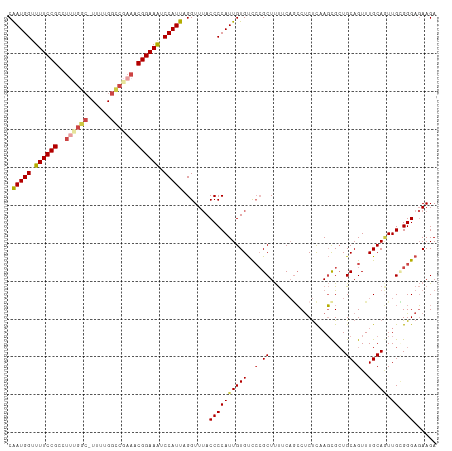
| Location | 18,901,773 – 18,901,881 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Shannon entropy | 0.29170 |
| G+C content | 0.51009 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -28.74 |
| Energy contribution | -29.80 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
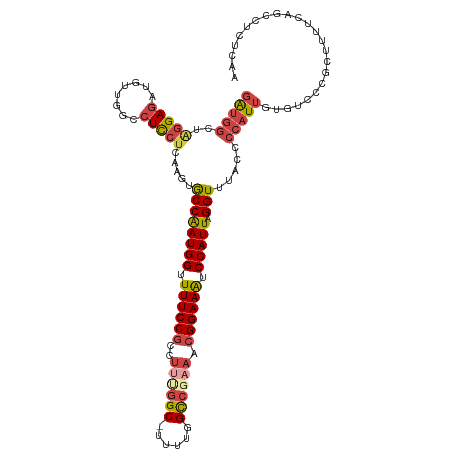
>dm3.chr2L 18901773 108 + 23011544 UUGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA-GC-------GGAAAACCAUUGGCACUUGAGGAGGCCAACAUCUCCUAACCAUC ..(((((((((((..((((((..((.((((.....)))).))...)))))))))))))....-..-------((....)).(((((.(((...))))))))..))))......... ( -35.10, z-score = -2.13, R) >droAna3.scaffold_12916 1914095 114 + 16180835 UUAGGGGCUGGGGGAGCCCAGCACAAUGGGGUUAACCUAAUGGAUUUCCGUUUGGACCAAAA-GCCAGAGGCGGAAAACCAUCGGUACUUGAGGAGGCCAGCCAUUCUCAGUUGC- ......((((((....))))))....((((.....))))((((.(((((((((((.......-.))))..))))))).))))..(((.(((((((((....)).))))))).)))- ( -42.80, z-score = -1.61, R) >droEre2.scaffold_4845 17213102 116 - 22589142 UUGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGACUUCCGUUUCGACCAAAAAGCCGAAGGCGGAAAACCAUUGGCACUUGAGGAGACCGACUUCUCCUGGCCAUC ......((((...((((((((..((.((((.....)))).))...))))))))..........(((((.((.......)).))))).....((((((......))))))))))... ( -36.60, z-score = -1.46, R) >droYak2.chr2R 5418098 116 + 21139217 UUGAGAGGAUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAAAGCCAAAGGCGGAAAACCAUUGGCACUUGAGAAGACCGACAACUCCGGGCCAUC .............((((((((..((.((((.....)))).))...)))))))).((((.....(((((.((.......)).)))))....(((..(.....)..)))..))))... ( -32.50, z-score = -0.83, R) >droSec1.super_7 2530567 115 + 3727775 UGGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA-GCCAAAGGCGGAAAACCAUUGGCACUUGAGGAGGCCAACAUCUCCUAACCAUC (((...(((((((..((((((..((.((((.....)))).))...)))))))))))))....-(((((.((.......)).))))).....((((((......))))))..))).. ( -42.10, z-score = -3.13, R) >droSim1.chr2L 18592018 115 + 22036055 UGGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA-GCCAAAGGCGGAAAACCAUUGGCACUUGAGGAGGCCAACAUCUCCUAACCAUC (((...(((((((..((((((..((.((((.....)))).))...)))))))))))))....-(((((.((.......)).))))).....((((((......))))))..))).. ( -42.10, z-score = -3.13, R) >consensus UUGAGAGGCUGAAAAGCGGGACACAAUGGGGUAAACCUAAUGGAUUUCCGUUUCGGCCAAAA_GCCAAAGGCGGAAAACCAUUGGCACUUGAGGAGGCCAACAUCUCCUAACCAUC (((((.(((((((..((((((..((.((((.....)))).))...))))))))))))).....(((((.((.......)).))))).)))))((((........))))........ (-28.74 = -29.80 + 1.06)
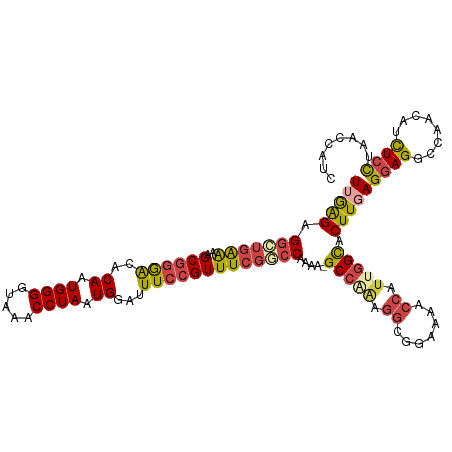
| Location | 18,901,773 – 18,901,881 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Shannon entropy | 0.29170 |
| G+C content | 0.51009 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -24.52 |
| Energy contribution | -25.75 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18901773 108 - 23011544 GAUGGUUAGGAGAUGUUGGCCUCCUCAAGUGCCAAUGGUUUUCC-------GC-UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCAA ((.((((.(((((.((.(((((.....)).)))(((((.(((((-------(.-((((....)))))))))).))))).((..(((......)))..))))))))))))))).... ( -29.50, z-score = -0.71, R) >droAna3.scaffold_12916 1914095 114 - 16180835 -GCAACUGAGAAUGGCUGGCCUCCUCAAGUACCGAUGGUUUUCCGCCUCUGGC-UUUUGGUCCAAACGGAAAUCCAUUAGGUUAACCCCAUUGUGCUGGGCUCCCCCAGCCCCUAA -((((.((((...((....))..))))...((((((((.((((((....((((-.....).)))..)))))).))))).)))........))))((((((....))))))...... ( -34.10, z-score = -0.52, R) >droEre2.scaffold_4845 17213102 116 + 22589142 GAUGGCCAGGAGAAGUCGGUCUCCUCAAGUGCCAAUGGUUUUCCGCCUUCGGCUUUUUGGUCGAAACGGAAGUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCAA ((.(((...(((((((.((.(....(((..((((((((.((((((..(((((((....))))))).)))))).))))).)))........))).).)).))))))).))))).... ( -40.20, z-score = -2.82, R) >droYak2.chr2R 5418098 116 - 21139217 GAUGGCCCGGAGUUGUCGGUCUUCUCAAGUGCCAAUGGUUUUCCGCCUUUGGCUUUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAUCCUCUCAA (((((.((((.....)))).......((((...(((((.((((((..(((((((....))))))).)))))).))))).((..(((......)))..)))))).)))))....... ( -33.50, z-score = -1.44, R) >droSec1.super_7 2530567 115 - 3727775 GAUGGUUAGGAGAUGUUGGCCUCCUCAAGUGCCAAUGGUUUUCCGCCUUUGGC-UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCCA ........(((((.((((((((.....)).)))(((((.((((((..((((((-.....)))))).)))))).))))).((..(((......)))..)).......))).))))). ( -39.00, z-score = -2.62, R) >droSim1.chr2L 18592018 115 - 22036055 GAUGGUUAGGAGAUGUUGGCCUCCUCAAGUGCCAAUGGUUUUCCGCCUUUGGC-UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCCA ........(((((.((((((((.....)).)))(((((.((((((..((((((-.....)))))).)))))).))))).((..(((......)))..)).......))).))))). ( -39.00, z-score = -2.62, R) >consensus GAUGGCUAGGAGAUGUUGGCCUCCUCAAGUGCCAAUGGUUUUCCGCCUUUGGC_UUUUGGCCGAAACGGAAAUCCAUUAGGUUUACCCCAUUGUGUCCCGCUUUUCAGCCUCUCAA (((((..(((((........))))).....((((((((.((((((..((((((......)))))).)))))).))))).))).....)))))........................ (-24.52 = -25.75 + 1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:55 2011