| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,798,832 – 1,798,929 |
| Length | 97 |
| Max. P | 0.983071 |

| Location | 1,798,832 – 1,798,929 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.46 |
| Shannon entropy | 0.47939 |
| G+C content | 0.43696 |
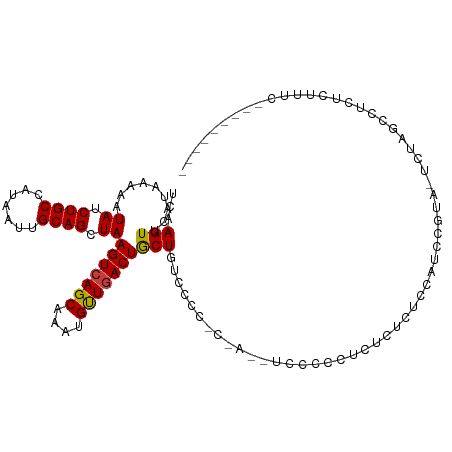
| Mean single sequence MFE | -16.29 |
| Consensus MFE | -12.00 |
| Energy contribution | -11.81 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
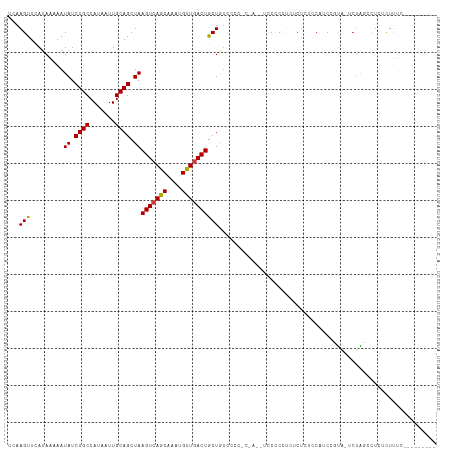
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
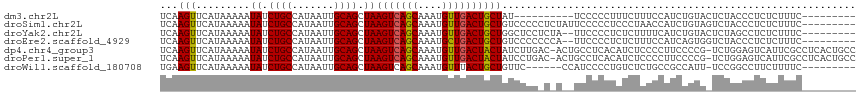
>dm3.chr2L 1798832 97 + 23011544 UCAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGUUGACUGCUAU----------UCCCCCUUUCUUUCCAUCUGUACUCUACCCUCUCUUUC--------- ...(((.((......((.((((.......)))).))(((((((....))))))).....----------..................)).)))..............--------- ( -12.70, z-score = -2.21, R) >droSim1.chr2L 1787441 107 + 22036055 UCAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGUUGACUGCUGUCCCCCUCUAUUCCCCCUCCCUAACCAUCUGUAGUCUACCCUCUCUUUC--------- .............................(((((..(((((((....))))))))))))................................................--------- ( -14.70, z-score = -2.88, R) >droYak2.chr2L 1774922 105 + 22324452 UCAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGUUGACUGCUGGCUCCUCUA--UUCCCCUCUCUUUUCAUCUGUACUCUAGCCUCUCUUUC--------- ...(((.........((.((((.......)))).))(((((((....))))))))))((((....((--(..................)))....))))........--------- ( -14.87, z-score = -0.88, R) >droEre2.scaffold_4929 1845091 105 + 26641161 UCAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGCUGACUGCUGUCCCCCCCA--UUCCCCUCUCUUUCCAUCAGUGGUCUACCCUCUCUUUC--------- .............................(((((..(((((((....)))))))))))).....(((--((................)))))...............--------- ( -18.49, z-score = -3.06, R) >dp4.chr4_group3 3183650 114 - 11692001 UCAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGUUGACUACUAUCUUGAC-ACUGCCUCACAUCUCCCCUUCCCCG-UCUGGAGUCAUUCGCCUCACUGCC (((((..........((.((((.......)))).))(((((((....))))))).....))))).-...((.......((((.........-...))))......))......... ( -19.02, z-score = -1.07, R) >droPer1.super_1 324622 114 - 10282868 UCAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGUUGACUACUAUCCUGAC-ACUGCCUCACAUCUCCCCUUCCCCG-UCUGGAGUCAUUCGCCUCACUGCC .............................((((...(((((((....)))))))....(((.(((-........................)-)).))).............)))). ( -17.86, z-score = -0.75, R) >droWil1.scaffold_180708 684837 100 - 12563649 UGAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGUUUACUGCUGUUC------CCAUCCCCUGUCUCUGCCGCCAUU-UCCGGCCUUCUUUUC--------- .((((...............(((..(((.((.((..((.(((...(((...((....))..------.)))...))).))..)).)).)))-...))))))).....--------- ( -16.36, z-score = -1.22, R) >consensus UCAAGUUCAUAAAAAUAUCUGCCAUAAUUGCAGCUAAGUCAGCAAAUGUUGACUGCUGUCCCC_C_A__UCCCCCUCUCUCUCCAUCCGUA_UCUAGCCUCUCUUUC_________ ...(((.........((.((((.......)))).))(((((((....))))))))))........................................................... (-12.00 = -11.81 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:14 2011