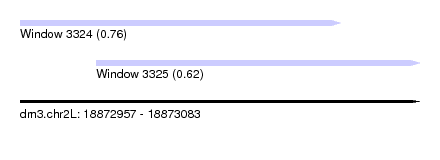
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,872,957 – 18,873,083 |
| Length | 126 |
| Max. P | 0.764928 |

| Location | 18,872,957 – 18,873,058 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.59 |
| Shannon entropy | 0.44276 |
| G+C content | 0.41191 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -15.69 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18872957 101 + 23011544 ----------------CCUUUGCUUGUUGUCUCUUCU--CGGCUGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGC ----------------.........((((........--))))......((((((.((((((...(((.(((((.(((((.....))))).))))).))).......)))))))))))) ( -18.70, z-score = -1.12, R) >droGri2.scaffold_15252 4863085 119 - 17193109 UGGUUGUUGUUUGUUUGCUGUUGCUUUUGCUCUCCAUGGCAGGCGGCUUGUGUAAAUAUUUGACCUGACGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGC ..(((((((..(((((((.((((((..((((......))))))))))....)))))))..))))..)))(((((.(((((.....))))).))))).........((((.....)))). ( -29.70, z-score = -0.58, R) >droMoj3.scaffold_6500 30555633 110 - 32352404 ---------UUGUUGCCUGUUGCCUUUUGCCUUCGCUGGCAGUUGGCUUGUGUAAAUAUUUGACCCGACGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGC ---------............(((..(((((......)))))..)))..((((((.((((((...(((((((((.(((((.....))))).))))).......)))))))))))))))) ( -31.41, z-score = -2.54, R) >droWil1.scaffold_180703 2212413 87 - 3946847 --------------------------------UGUGUGGCAGGCGGCUUAUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGC --------------------------------.(((..(.(.(((((....(((((.(((......)))(((((.(((((.....))))).))))))))))..)))))...).)..))) ( -18.40, z-score = 0.46, R) >droAna3.scaffold_12916 1889682 93 + 16180835 --------------------------CCUCUCCCACUGCCACUGCCCCUCUGCAAAUAUUUGACCUGGUGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGC --------------------------.........................((((.((((((.......(((((.(((((.....))))).)))))...........)))))))))).. ( -14.87, z-score = -0.26, R) >droEre2.scaffold_4845 17185019 101 - 22589142 ----------------CCUUUGCCUGUUUUCCCUCCU--CGGCUGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGCCGCAAAUAUUGCGC ----------------.....(((.............--.)))......((((((.((((((...(((.(((((.(((((.....))))).))))).))).......)))))))))))) ( -19.64, z-score = -1.97, R) >droSec1.super_7 2500084 101 + 3727775 ----------------CCUUUGCUUGUUGUCUCUCCU--CGGCCGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGC ----------------.........((((........--))))......((((((.((((((...(((.(((((.(((((.....))))).))))).))).......)))))))))))) ( -18.30, z-score = -0.97, R) >droSim1.chr2L 18563245 101 + 22036055 ----------------CCUUUGCUUGUUGUCUCUCCC--CGGCUGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGC ----------------.........((((........--))))......((((((.((((((...(((.(((((.(((((.....))))).))))).))).......)))))))))))) ( -18.20, z-score = -1.18, R) >consensus ________________CCUUUGCCUGUUGUCUCUCCU__CAGCUGUCUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGC .................................................((((((.((((((...(((.(((((.(((((.....))))).))))).))).......)))))))))))) (-15.69 = -15.60 + -0.09)
| Location | 18,872,981 – 18,873,083 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 80.27 |
| Shannon entropy | 0.41250 |
| G+C content | 0.39448 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -18.87 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18872981 102 + 23011544 ---CUGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAC--GACAACACCAAAGGGUGCA-------------- ---.((((((((((((.((((((...(((.(((((.(((((.....))))).))))).))).......))))))))))))))).))--)....((((....))))..-------------- ( -26.50, z-score = -1.59, R) >droGri2.scaffold_15252 4863124 116 - 17193109 CAGGCGGCUUGUGUAAAUAUUUGACCUGACGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACACUGAGACACUCGCAGAAAAGCAAGAGCAGCU----- ..(((.((((((((((.((((((....((((((((.(((((.....))))).))))).......))).)))))))))))).....(((.(.....).)))........)))).)))----- ( -27.91, z-score = 0.36, R) >droMoj3.scaffold_6500 30555663 118 - 32352404 CAGUUGGCUUGUGUAAAUAUUUGACCCGACGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAC--GAACAGACCAAGCACAGCAC-CUCAUCUCAUCU ..(((((((((.((...(.((((.......(((((.(((((.....))))).)))))....((((.((((.....))))))))..)--))).).))))))).))))..-............ ( -28.20, z-score = -1.40, R) >droVir3.scaffold_12963 9491383 113 + 20206255 CAGGCGGCUUGUGUAAAUAUUUGACCUGACGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAU--GAACAGGC-AAGCAAAGCGGACCCACCU----- ..(((.((((.(((......(((.((((..(((((.(((((.....))))).)))))....((((.((((.....))))))))...--...)))))-))))))))).).)).....----- ( -29.50, z-score = -0.64, R) >droWil1.scaffold_180703 2212423 84 - 3946847 ---GCGGCUUAUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAUG---------------------------------- ---(((((....)).((((((((...(((.(((((.(((((.....))))).))))).))).......)))))))).))).......---------------------------------- ( -16.30, z-score = -0.13, R) >droPer1.super_1 3704123 104 + 10282868 CGGUGCUCUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAU--GACAACAC-AAAAGGUGCA-------------- ..(..((.((((((............(((.(((((.(((((.....))))).))))).)))((((.((((.....))))))))...--....))))-))..))..).-------------- ( -24.60, z-score = -0.49, R) >dp4.chr4_group3 2233545 104 + 11692001 CGGUGCUCUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAU--GACAACAC-AAAAGGUGCA-------------- ..(..((.((((((............(((.(((((.(((((.....))))).))))).)))((((.((((.....))))))))...--....))))-))..))..).-------------- ( -24.60, z-score = -0.49, R) >droAna3.scaffold_12916 1889703 97 + 16180835 --------CUCUGCAAAUAUUUGACCUGGUGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCGAACAC--GACAAGACGAGAGGUCUUG-------------- --------..............(((((...(((((.(((((.....))))).)))))....(((((((((.....)))(((....)--).)..)))))))))))...-------------- ( -22.40, z-score = -0.61, R) >droEre2.scaffold_4845 17185043 99 - 22589142 ---CUGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGCCGCAAAUAUUGCGCAAACAC--GACA---CCAAAGGGUGCA-------------- ---..(((((((((((.((((((...(((.(((((.(((((.....))))).))))).))).......))))))))))))))).))--(.((---((....))))).-------------- ( -27.10, z-score = -2.06, R) >droYak2.chr2R 5388864 102 + 21139217 ---CUGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAC--GACAACGCCAAAGGGUGCA-------------- ---.((((((((((((.((((((...(((.(((((.(((((.....))))).))))).))).......))))))))))))))).))--)....((((....))))..-------------- ( -26.10, z-score = -1.17, R) >droSec1.super_7 2500108 102 + 3727775 ---CCGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAC--GACAACACCAAAGGGUGCA-------------- ---.((((((((((((.((((((...(((.(((((.(((((.....))))).))))).))).......))))))))))))))).))--)....((((....))))..-------------- ( -28.50, z-score = -2.24, R) >droSim1.chr2L 18563269 102 + 22036055 ---CUGUUUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUCUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAC--GACAACACCAAAGGGUGCA-------------- ---.((((((((((((.((((((...(((.(((((.(((((.....))))).))))).))).......))))))))))))))).))--)....((((....))))..-------------- ( -26.50, z-score = -1.59, R) >consensus ___CUGUCUUGUGUAAAUAUUUGACCUGAUGCGUGUGAACUUUAACAGUUUUUAUGCUUUAUUUGUCGCAAAUAUUGCGCAAACAC__GACAACACCAAAGGGUGCA______________ ........((((((((.((((((...(((.(((((.(((((.....))))).))))).))).......))))))))))))))....................................... (-18.87 = -18.73 + -0.14)
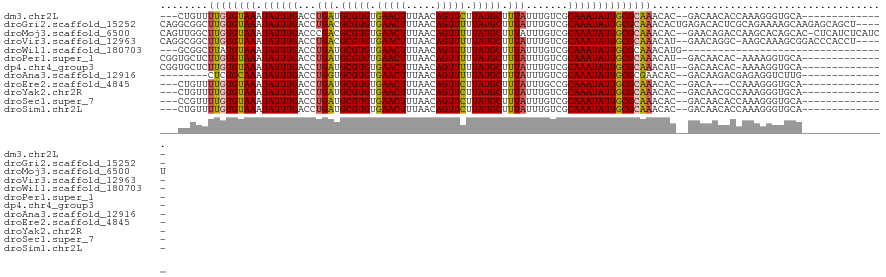

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:49 2011