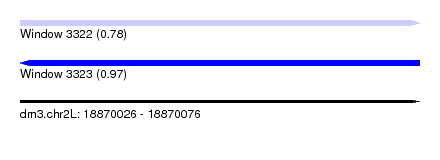
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,870,026 – 18,870,076 |
| Length | 50 |
| Max. P | 0.973782 |

| Location | 18,870,026 – 18,870,076 |
|---|---|
| Length | 50 |
| Sequences | 7 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Shannon entropy | 0.30214 |
| G+C content | 0.42642 |
| Mean single sequence MFE | -14.37 |
| Consensus MFE | -10.79 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
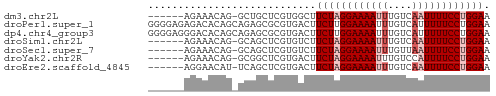
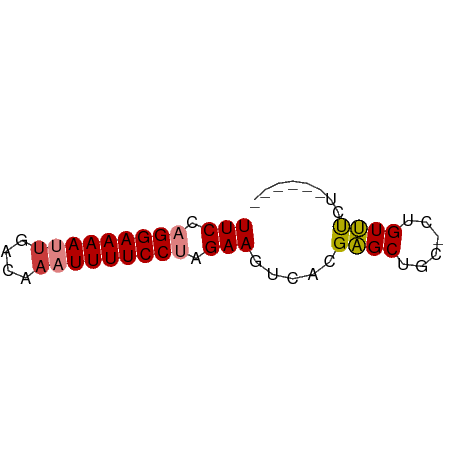
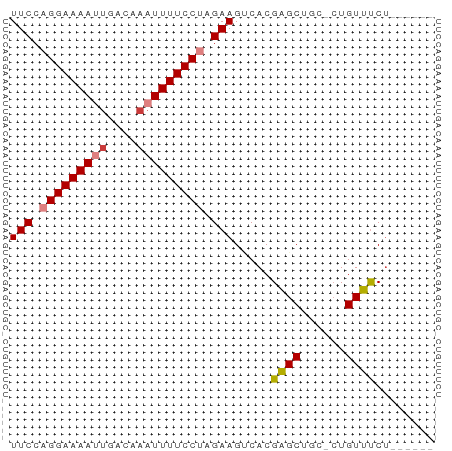
>dm3.chr2L 18870026 50 + 23011544 ------AGAAACAG-GCUGCUCGUGGCUUCUAGGAAAAUUUGUCAAUUUUCCUGGAA ------.......(-((((....)))))((((((((((((....)))))))))))). ( -16.10, z-score = -2.52, R) >droPer1.super_1 3700207 57 + 10282868 GGGGAGAGACACAGCAGAGCGCGUGACUUCUUGGAAAAUUUGUCAUUUUUCCUGGAA ((((((...(((.((.....))))).))))))((((((........))))))..... ( -12.30, z-score = 0.26, R) >dp4.chr4_group3 2229606 57 + 11692001 GGGGAGGGACACAGCAGAGCGCGUGACUUCUUGGAAAAUUUGUCAUUUUUCCUGGAA .((((((..(((.((.....))))).))))))((((((........))))))..... ( -14.60, z-score = -0.52, R) >droSim1.chr2L 18560318 50 + 22036055 ------AGAAACAG-GCAGCUCGUGUCUUCUAGGAAAAUUUGUCAAUUUUCCUGGAA ------......((-(((.....)))))((((((((((((....)))))))))))). ( -15.70, z-score = -2.78, R) >droSec1.super_7 2497153 50 + 3727775 ------AGAAACAG-GCAGCUCGUGUCUUCUAGGAAAAUUUGUUAAUUUUCCUGGAA ------......((-(((.....)))))((((((((((((....)))))))))))). ( -16.40, z-score = -3.27, R) >droYak2.chr2R 5385967 50 + 21139217 ------AGAAACAG-GCGGCUCGUGACUUCUAGGAAAAUUUGUCCAUUUUCCUGGAA ------.((..(..-..)..))......(((((((((((......))))))))))). ( -12.50, z-score = -0.84, R) >droEre2.scaffold_4845 17182750 50 - 22589142 ------AGGAACAU-UCAGCUCGUGACUUCUAGGAAAAUUUGUCAAUUUUCCUGGAA ------........-.............((((((((((((....)))))))))))). ( -13.00, z-score = -1.81, R) >consensus ______AGAAACAG_GCAGCUCGUGACUUCUAGGAAAAUUUGUCAAUUUUCCUGGAA ............................((((((((((((....)))))))))))). (-10.79 = -11.50 + 0.71)
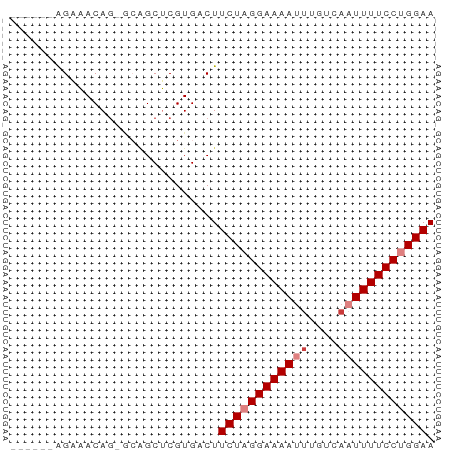
| Location | 18,870,026 – 18,870,076 |
|---|---|
| Length | 50 |
| Sequences | 7 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 83.23 |
| Shannon entropy | 0.30214 |
| G+C content | 0.42642 |
| Mean single sequence MFE | -10.96 |
| Consensus MFE | -10.59 |
| Energy contribution | -10.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
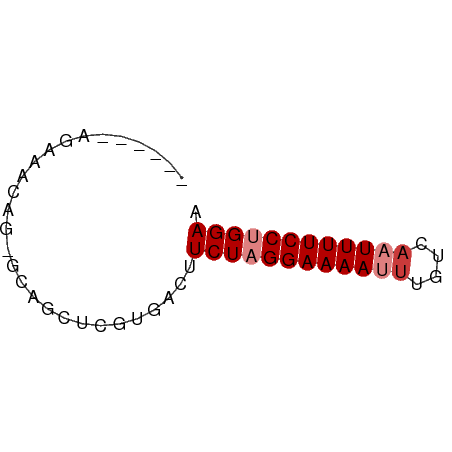
>dm3.chr2L 18870026 50 - 23011544 UUCCAGGAAAAUUGACAAAUUUUCCUAGAAGCCACGAGCAGC-CUGUUUCU------ (((.(((((((((....))))))))).)))((.....))...-........------ ( -11.50, z-score = -2.03, R) >droPer1.super_1 3700207 57 - 10282868 UUCCAGGAAAAAUGACAAAUUUUCCAAGAAGUCACGCGCUCUGCUGUGUCUCUCCCC .....((((((........)))))).(((.(.(((((.....)).))).)))).... ( -10.20, z-score = -0.94, R) >dp4.chr4_group3 2229606 57 - 11692001 UUCCAGGAAAAAUGACAAAUUUUCCAAGAAGUCACGCGCUCUGCUGUGUCCCUCCCC (((..((((((........))))))..)))(.(((((.....)).))).)....... ( -9.00, z-score = -0.62, R) >droSim1.chr2L 18560318 50 - 22036055 UUCCAGGAAAAUUGACAAAUUUUCCUAGAAGACACGAGCUGC-CUGUUUCU------ (((.(((((((((....))))))))).))).....((((...-..))))..------ ( -11.50, z-score = -1.97, R) >droSec1.super_7 2497153 50 - 3727775 UUCCAGGAAAAUUAACAAAUUUUCCUAGAAGACACGAGCUGC-CUGUUUCU------ (((.(((((((((....))))))))).))).....((((...-..))))..------ ( -10.60, z-score = -2.09, R) >droYak2.chr2R 5385967 50 - 21139217 UUCCAGGAAAAUGGACAAAUUUUCCUAGAAGUCACGAGCCGC-CUGUUUCU------ (((.((((((((......)))))))).)))............-........------ ( -10.10, z-score = -0.75, R) >droEre2.scaffold_4845 17182750 50 + 22589142 UUCCAGGAAAAUUGACAAAUUUUCCUAGAAGUCACGAGCUGA-AUGUUCCU------ (((.(((((((((....))))))))).))).....((((...-..))))..------ ( -13.80, z-score = -3.04, R) >consensus UUCCAGGAAAAUUGACAAAUUUUCCUAGAAGUCACGAGCUGC_CUGUUUCU______ (((.(((((((((....))))))))).))).....((((......))))........ (-10.59 = -10.77 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:47 2011