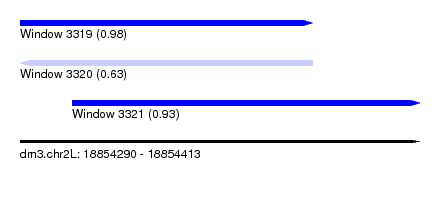
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,854,290 – 18,854,413 |
| Length | 123 |
| Max. P | 0.981863 |

| Location | 18,854,290 – 18,854,380 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Shannon entropy | 0.42488 |
| G+C content | 0.51723 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.44 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
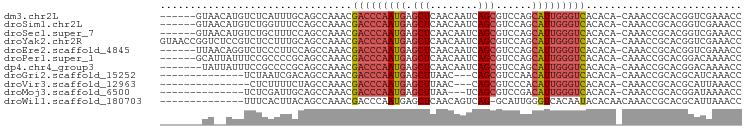
>dm3.chr2L 18854290 90 + 23011544 ------GUAACAUGUCUCAUUUGCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA-CAAACCGCACGGUCGAAACC ------.......((.((..............(((((((((.(((........)))......))))))))).....-...(((....))).)).)). ( -19.80, z-score = -1.08, R) >droSim1.chr2L 18543772 90 + 22036055 ------GUAACAUGUCUGGUUUCCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA-CAAACCGCACGGUCGAAACC ------...........((((((.........(((((((((.(((........)))......))))))))).....-...(((....))).)))))) ( -24.90, z-score = -2.30, R) >droSec1.super_7 2480697 90 + 3727775 ------GUAACAUGUCUGCUUUCCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA-CAAACCGCACGGUCGAAACC ------......((.(((.....))).))...(((((((((.(((........)))......))))))))).....-...(((....)))....... ( -20.30, z-score = -1.18, R) >droYak2.chr2R 5369924 96 + 21139217 GUAACCGGUCUCCGUCUCCUUUGCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA-CAAACCGCACGGUCGAAACC ...(((((.((.((.......)).))))....(((((((((.(((........)))......))))))))).....-..........)))....... ( -22.20, z-score = -0.96, R) >droEre2.scaffold_4845 17167084 90 - 22589142 ------UUAACAGGUCUCCCUUCCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA-CAAACCGCACGGUCGAAACC ------......(((.((..............(((((((((.(((........)))......))))))))).....-...(((....))).)).))) ( -21.60, z-score = -2.33, R) >droPer1.super_1 3683113 90 + 10282868 ------GCAUUAUUUCCGCCCCGCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA-CAAACCGCACGGACAAAACC ------........(((((......)).....(((((((((.(((........)))......))))))))).....-..........)))....... ( -20.30, z-score = -2.15, R) >dp4.chr4_group3 2212488 89 + 11692001 -------UAUUAUUUCCGCCCCGCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA-CAAACCGCACGGACAAAACC -------.......(((((......)).....(((((((((.(((........)))......))))))))).....-..........)))....... ( -20.30, z-score = -2.64, R) >droGri2.scaffold_15252 4839021 79 - 17193109 --------------UCUAAUCGACAGCCAAACGACCCAAUGAGCUUAAC---CAGCGUCCAACAUUGGGUCACACA-CAAACCGCACGCAUCAAACC --------------..................(((((((((.(((....---.)))......))))))))).....-.................... ( -15.80, z-score = -2.62, R) >droVir3.scaffold_12963 9471269 78 + 20206255 ---------------CUCUUUUCUAGCCAAACGACCCAAUGAGCUUAAC---CAGCGUCCCACAUUGGGUCACACA-CAAACCGCACGCAUUAAACC ---------------.................(((((((((.(((....---.)))......))))))))).....-.................... ( -15.80, z-score = -2.81, R) >droMoj3.scaffold_6500 30536773 79 - 32352404 --------------UCUCGAUUGCAGCCAAACGACCCAAUGAGCUUAA---UCAGCGUCCGACAUUGGGUCACACA-CAAACCGCACGGAUAAAACC --------------((.((..(((.(.....)(((((((((.(((...---..)))......))))))))).....-......)))))))....... ( -17.20, z-score = -1.46, R) >droWil1.scaffold_180703 2188831 82 - 3946847 --------------UUUCACUUACAGCCAAACGACCCAAUGAGCUCAACAGUCAG-GCAUUGGGUCACAAUACACAACAAACCGCACGCAUUAAACC --------------..................(((((((((.(((....)))...-.)))))))))............................... ( -15.60, z-score = -2.43, R) >consensus _______UA__AUGUCUCCUUUGCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACACA_CAAACCGCACGGACGAAACC ................................(((((((((.(((........)))......))))))))).......................... (-13.96 = -14.44 + 0.47)
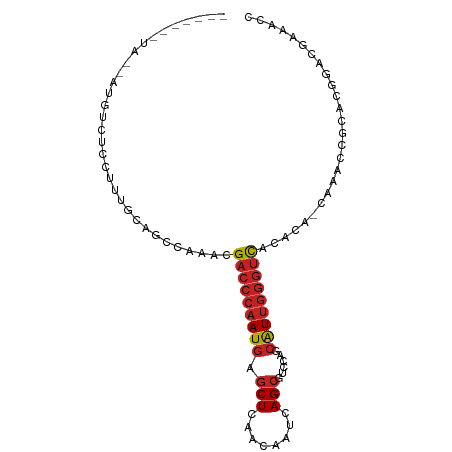
| Location | 18,854,290 – 18,854,380 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Shannon entropy | 0.42488 |
| G+C content | 0.51723 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -14.55 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18854290 90 - 23011544 GGUUUCGACCGUGCGGUUUG-UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGCAAAUGAGACAUGUUAC------ .((((((....(((((((..-....((((((((((((.((((.....)))).)))..)))))))))...)))))))..)))))).......------ ( -32.50, z-score = -2.41, R) >droSim1.chr2L 18543772 90 - 22036055 GGUUUCGACCGUGCGGUUUG-UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGGAAACCAGACAUGUUAC------ ((((((((((....))))..-.....(((((((((((.((((.....)))).)))..)))))))).........))))))...........------ ( -30.70, z-score = -1.57, R) >droSec1.super_7 2480697 90 - 3727775 GGUUUCGACCGUGCGGUUUG-UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGGAAAGCAGACAUGUUAC------ ......((((....))))..-.....(((((((((((.((((.....)))).)))..))))))))(((((.((....))))))).......------ ( -29.40, z-score = -1.12, R) >droYak2.chr2R 5369924 96 - 21139217 GGUUUCGACCGUGCGGUUUG-UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGCAAAGGAGACGGAGACCGGUUAC ((((((.....(((((((..-....((((((((((((.((((.....)))).)))..)))))))))...)))))))..(....).))))))...... ( -37.00, z-score = -2.36, R) >droEre2.scaffold_4845 17167084 90 + 22589142 GGUUUCGACCGUGCGGUUUG-UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGGAAGGGAGACCUGUUAA------ ((((((..((.(.(((((..-....((((((((((((.((((.....)))).)))..)))))))))...))))).).))))))))......------ ( -32.10, z-score = -2.02, R) >droPer1.super_1 3683113 90 - 10282868 GGUUUUGUCCGUGCGGUUUG-UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGCGGGGCGGAAAUAAUGC------ ..((((((((.(((((((..-....((((((((((((.((((.....)))).)))..)))))))))...))))))))))))))).......------ ( -35.70, z-score = -3.09, R) >dp4.chr4_group3 2212488 89 - 11692001 GGUUUUGUCCGUGCGGUUUG-UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGCGGGGCGGAAAUAAUA------- ..((((((((.(((((((..-....((((((((((((.((((.....)))).)))..)))))))))...)))))))))))))))......------- ( -35.70, z-score = -3.34, R) >droGri2.scaffold_15252 4839021 79 + 17193109 GGUUUGAUGCGUGCGGUUUG-UGUGUGACCCAAUGUUGGACGCUG---GUUAAGCUCAUUGGGUCGUUUGGCUGUCGAUUAGA-------------- ..((((((.((.((((((..-....((((((((((...(((....---))).....))))))))))...))))))))))))))-------------- ( -24.20, z-score = -1.33, R) >droVir3.scaffold_12963 9471269 78 - 20206255 GGUUUAAUGCGUGCGGUUUG-UGUGUGACCCAAUGUGGGACGCUG---GUUAAGCUCAUUGGGUCGUUUGGCUAGAAAAGAG--------------- (((..((((((..(.....)-..)))(((((((((.(.(((....---)))...).))))))))))))..))).........--------------- ( -22.00, z-score = -1.00, R) >droMoj3.scaffold_6500 30536773 79 + 32352404 GGUUUUAUCCGUGCGGUUUG-UGUGUGACCCAAUGUCGGACGCUGA---UUAAGCUCAUUGGGUCGUUUGGCUGCAAUCGAGA-------------- ((......)).(((((((..-....((((((((((......(((..---...))).))))))))))...))))))).......-------------- ( -25.10, z-score = -1.24, R) >droWil1.scaffold_180703 2188831 82 + 3946847 GGUUUAAUGCGUGCGGUUUGUUGUGUAUUGUGACCCAAUGC-CUGACUGUUGAGCUCAUUGGGUCGUUUGGCUGUAAGUGAAA-------------- ........((.(((((((............((((((((((.-((........))..))))))))))...))))))).))....-------------- ( -18.96, z-score = -0.04, R) >consensus GGUUUCGUCCGUGCGGUUUG_UGUGUGACCCAAUGCUGGACGCUGAUUGUUGAGCUCAUUGGGUCGUUUGGCUGCAAAGGAGACAU__UA_______ .............(((((.......((((((((((......(((........))).))))))))))...)))))....................... (-14.55 = -14.95 + 0.39)


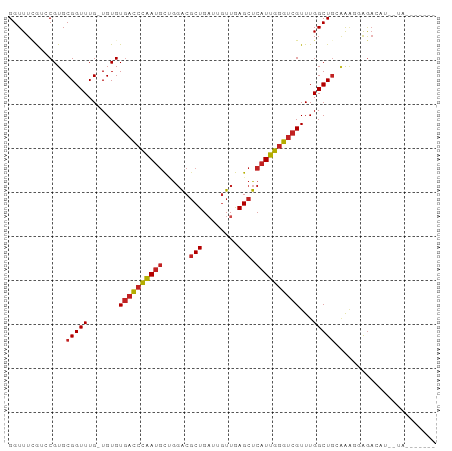
| Location | 18,854,306 – 18,854,413 |
|---|---|
| Length | 107 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Shannon entropy | 0.38699 |
| G+C content | 0.52278 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
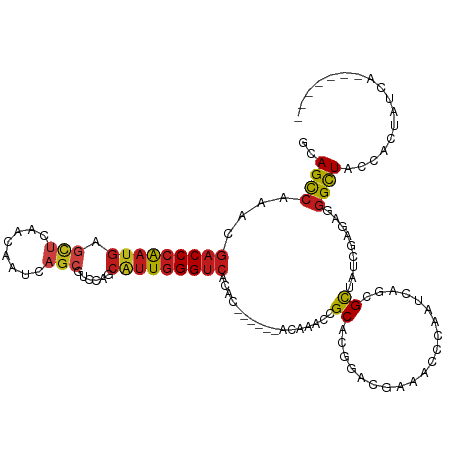
>dm3.chr2L 18854306 107 + 23011544 GCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC------ACAAACCGCACGGUCGAAACCCAAUCAGCGCUAUCGAGAGGGCUACCACUAUCA------- ..((((....(((((((((.(((........)))......)))))))))....------......(((..(((....)))......)))..........))))..........------- ( -25.60, z-score = -1.32, R) >droSim1.chr2L 18543788 107 + 22036055 CCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC------ACAAACCGCACGGUCGAAACCCAAUCAACGCUAUCGAGAGGGCUACCACUAUCA------- ..((((....(((((((((.(((........)))......)))))))))....------..........(..((((................))))..)))))..........------- ( -25.29, z-score = -1.91, R) >droSec1.super_7 2480713 107 + 3727775 CCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC------ACAAACCGCACGGUCGAAACCCAAUCAGCGCUAUCGAGAGGGCUACCACUAUCA------- ..((((....(((((((((.(((........)))......)))))))))....------......(((..(((....)))......)))..........))))..........------- ( -25.60, z-score = -1.57, R) >droYak2.chr2R 5369946 107 + 21139217 GCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC------ACAAACCGCACGGUCGAAACCCAAUCAGCGCUAUAGAGAGGGCUACCACUAUCA------- ..((((....(((((((((.(((........)))......)))))))))....------......(((..(((....)))......)))..........))))..........------- ( -25.60, z-score = -1.51, R) >droEre2.scaffold_4845 17167100 107 - 22589142 CCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC------ACAAACCGCACGGUCGAAACCCAAUCAGCGCUACAGAGAGGGCUACCACUAUCA------- ..((((....(((((((((.(((........)))......)))))))))....------......(((..(((....)))......)))..........))))..........------- ( -25.60, z-score = -1.70, R) >droAna3.scaffold_12943 4648167 107 + 5039921 AUAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCGUUGGGUCACAC------ACAAACCGCACGGUCGAAACCCAAUCAGCGCUACAGAGAGGGCUACCACUAUCA------- .(((((....(((((((((.(((........)))......)))))))))....------......(((..(((....)))......)))..........))))).........------- ( -26.30, z-score = -1.51, R) >droPer1.super_1 3683129 107 + 10282868 GCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC------ACAAACCGCACGGACAAAACCCAAUCAACGUUAUCGAGAGGGCUAUCACUAUCA------- ..((((.((((((((((((.(((........)))......)))))))))....------.....((....))................)))..(....)))))..........------- ( -25.60, z-score = -2.51, R) >dp4.chr4_group3 2212503 107 + 11692001 GCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC------ACAAACCGCACGGACAAAACCCAAUCAACGUUAUCGAGAGGGCUAUCACUAUCA------- ..((((.((((((((((((.(((........)))......)))))))))....------.....((....))................)))..(....)))))..........------- ( -25.60, z-score = -2.51, R) >droGri2.scaffold_15252 4839029 111 - 17193109 ACAGCCAAACGACCCAAUGAGCUUAAC---CAGCGUCCAACAUUGGGUCACAC------ACAAACCGCACGCAUCAAACCCAAUCAGCGUUAUCGCGAGGGCUAUCAUUACCAGCCGGUU ..((((....(((((((((.(((....---.)))......)))))))))....------......(((((((..............))))....)))..))))......(((....))). ( -29.14, z-score = -2.72, R) >droVir3.scaffold_12963 9471276 111 + 20206255 CUAGCCAAACGACCCAAUGAGCUUAAC---CAGCGUCCCACAUUGGGUCACAC------ACAAACCGCACGCAUUAAACCCAAUCAGCGUUAUCGCGAGGGCUAUCACUACCAACCGGUU .(((((....(((((((((.(((....---.)))......)))))))))....------......(((((((..............))))....)))..))))).....(((....))). ( -30.14, z-score = -3.22, R) >droMoj3.scaffold_6500 30536781 111 - 32352404 GCAGCCAAACGACCCAAUGAGCUUAAU---CAGCGUCCGACAUUGGGUCACAC------ACAAACCGCACGGAUAAAACCGAAUCAGCGUUAUCGCGAGGGCUAUCACUACCAGCCGGUU ..........(((((((((.(((....---.)))......)))))))))....------...((((...(((......))).....(((....)))...((((.........)))))))) ( -30.00, z-score = -1.52, R) >droWil1.scaffold_180703 2188839 107 - 3946847 ACAGCCAAACGACCCAAUGAGCUCAAC------AGUCAGGCAUUGGGUCACAAUACACAACAAACCGCACGCAUUAAACCCAAUCAACGCUAUCGAGAGGGCUACCACUAUCA------- ..((((....(((((((((.(((....------)))....)))))))))............................................(....)))))..........------- ( -22.00, z-score = -1.96, R) >anoGam1.chr3R 44449551 102 + 53272125 GUAGUGAAACGUCCCGACCAAUCGUACAACCAGCGACCUACCCUAGGUAAUAU-------------GCA-----GAAGUCGAAUCACCGCAAUCGGCAGGGUUACCACUAUCAACCAAUC ((((((....((..(((....))).))((((.((((((((...)))))....)-------------)).-----...(((((..........)))))..))))..))))))......... ( -19.30, z-score = -0.10, R) >consensus GCAGCCAAACGACCCAAUGAGCUCAACAAUCAGCGUCCAGCAUUGGGUCACAC______ACAAACCGCACGGACGAAACCCAAUCAGCGCUAUCGAGAGGGCUACCACUAUCA_______ ..((((....(((((((((.(((........)))......))))))))).................((....................)).........))))................. (-17.52 = -17.87 + 0.35)
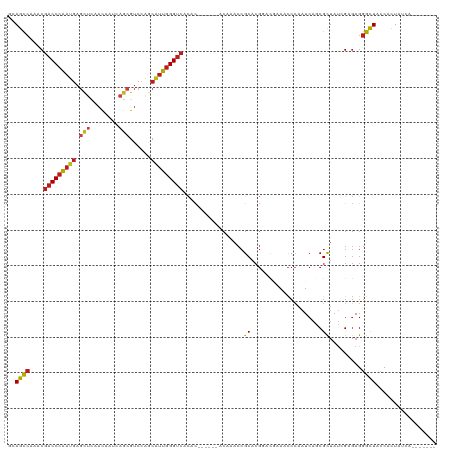
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:46 2011