
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,842,879 – 18,842,938 |
| Length | 59 |
| Max. P | 0.842224 |

| Location | 18,842,879 – 18,842,938 |
|---|---|
| Length | 59 |
| Sequences | 13 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 64.04 |
| Shannon entropy | 0.78518 |
| G+C content | 0.49473 |
| Mean single sequence MFE | -14.20 |
| Consensus MFE | -5.81 |
| Energy contribution | -5.76 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.90 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
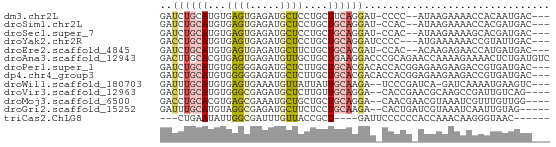
>dm3.chr2L 18842879 59 - 23011544 GAUCUGCAUGUGAGUGAGAUGCUCCUGCUUCAGGAU-CCCC--AUAAGAAAACCACAAUGAC--- ((((((((.(.((((.....))))))))....))))-)...--...................--- ( -10.10, z-score = 0.01, R) >droSim1.chr2L 18532225 59 - 22036055 GAUCUGCAUGUGAGUGAGAUGCUCCUGCUGCAGGAU-CCAC--AUAAGAAAACCACGAUGAC--- ((((((((.((((((.....))))..)))))).)))-)...--...................--- ( -12.20, z-score = -0.41, R) >droSec1.super_7 2469334 59 - 3727775 GAUCUGCAUGUGAGUGAGAUGCUCCUGCUGCAGGAU-CCAC--AUAAGAAAAGCACGAUGAC--- ((((((((.((((((.....))))..)))))).)))-)...--...................--- ( -12.20, z-score = 0.13, R) >droYak2.chr2R 5357579 59 - 21139217 GACCUGCAUGUGAGUGAGAUGCUCCUGCUGCAGGAUCCCC---AUGAAAAAACCGUAUUGAC--- ..((((((.((((((.....))))..))))))))......---...................--- ( -14.20, z-score = -2.41, R) >droEre2.scaffold_4845 17155690 59 + 22589142 GAUCUGCAUGUGAGUGAGAUGCUUCUGCUGCACGAU-CCAC--ACAAGAGAACCAUGAUGAC--- ((((((((.((((((.....))))..)))))).)))-)...--...................--- ( -10.60, z-score = 0.72, R) >droAna3.scaffold_12943 4636439 65 - 5039921 GACUUGCACGUGAGUGAGAUGUUGCUGCUGAAGGACCCGCAGAACCAAAAGAAAACUCUGAUGUC (((...(((....)))....(((.((((.(......).))))))).................))) ( -12.20, z-score = -0.22, R) >droPer1.super_1 3669736 62 - 10282868 GAUCUGCAUGUGGGGGAGAUGCUCUUGCUGCACGACACCACGGAGAAGAAGACCGUGAUGAC--- ..((((((.(..((((.....))))..))))).))((.(((((.........))))).))..--- ( -19.50, z-score = -1.97, R) >dp4.chr4_group3 2198548 62 - 11692001 GAUCUGCAUGUGGGGGAGAUGCUCUUGCUGCACGACACCACGGAGAAGAAGACCGUGAUGAC--- ..((((((.(..((((.....))))..))))).))((.(((((.........))))).))..--- ( -19.50, z-score = -1.97, R) >droWil1.scaffold_180703 2173903 59 + 3946847 GAUUUGCAUGUGAGUGAAAUGUUAUUAUUGCAAGA--UCCCGAUCA-GAUCAAAAUGAAGUC--- ...(((((....(((((....)))))..)))))((--((.......-))))...........--- ( -8.30, z-score = 0.04, R) >droVir3.scaffold_12963 10038741 59 + 20206255 GACUUGCAUGUGGGCGAGAUGCUCUUGUUGCAGGA--CACCGAACGCAAGCCGAUUGUCAG---- (((..((.((((..((.(.((.(((((...)))))--)))))..)))).)).....)))..---- ( -15.80, z-score = 0.08, R) >droMoj3.scaffold_6500 29501090 59 + 32352404 GACCUGCACGUGAGCGAAAUGCUGCUGCUGCAGGA--CAACGAACGUAAAUCGUUUGUUGG---- ..((((((.((.((((......)))))))))))).--(((((((((.....))))))))).---- ( -25.70, z-score = -4.58, R) >droGri2.scaffold_15252 3895462 58 + 17193109 GAUUUGCAUGUAGGCGAGAUGCUUCUCCUGCAAGA--CACUGAUCGUAAAUCAAUUGUAG----- (((((((.((((((.(((....))).)))))).((--......)))))))))........----- ( -16.40, z-score = -2.10, R) >triCas2.ChLG8 8907433 52 + 15773733 ---CUGAAUAUUGGCGAUUUGUUACCGCU----GAUUCCCCCCACCAAACAAGGGUAAC------ ---......((..(((.........))).----.))....(((.........)))....------ ( -7.90, z-score = 0.28, R) >consensus GAUCUGCAUGUGAGUGAGAUGCUCCUGCUGCAGGAU_CCCCG_ACAAGAAAACCAUGAUGAC___ ..((((((...((((.....))))....))))))............................... ( -5.81 = -5.76 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:40 2011