
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,782,036 – 1,782,133 |
| Length | 97 |
| Max. P | 0.828346 |

| Location | 1,782,036 – 1,782,133 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.28 |
| Shannon entropy | 0.57276 |
| G+C content | 0.50332 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -14.72 |
| Energy contribution | -14.74 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.828346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
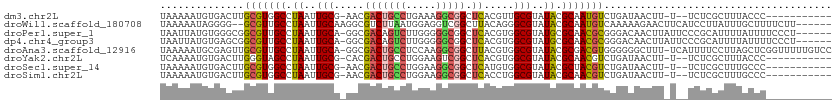
>dm3.chr2L 1782036 97 + 23011544 UAAAAAUGUGACUUGCGUGGCCUAAUUGCG-AACGACUGCCUGAAAGGCGGCUCACGUUGCGUAUACGCAAUGUCUGAUAACUU-U--UCUCGCUUUACCC----------- .......((.((....)).))......(((-(..(((((((.....))))).))((((((((....))))))))..........-.--..)))).......----------- ( -26.00, z-score = -1.87, R) >droWil1.scaffold_180708 656608 104 - 12563649 UAAAAAUAGGGG--GCGUUGCCUAAUUGCAAGGCGUCUUAAUGGAGGUCGGCUUACAGGGCGUAUACGCAAUGUCAAAAAGAACUUCAUCCUUAUUUGCUUUUCUU------ ...(((((((((--((.((((......)))).)).......(((((.((.....(((..(((....)))..)))......)).)))))))))))))).........------ ( -28.70, z-score = -1.65, R) >droPer1.super_1 306701 105 - 10282868 UAAUUAUGUGGGCGGCGUUGCCUAAUUGCA-GGCGACAGUCUUGGGGGCGGCUCACGUGGCGUAUGCGCAACGCGGGACAACUUAUUCCCGCAUUUUAUUUUCCCU------ ....(((((((((.(((((((((......)-))))))..((....)))).)))))))))(((....)))...((((((........))))))..............------ ( -41.80, z-score = -2.26, R) >dp4.chr4_group3 3165438 105 - 11692001 UAAUUAUGUGAGCGGCGUUGCCUAAUUGCA-GGCGACAGUCUUGGGGGCGGCUCACGUGGCGUAUGCGCAACGCGGGACAACUUAUUCCCGCAUUUUAUUUUCCCU------ ....(((((((((.(((((((((......)-))))))..((....)))).)))))))))(((....)))...((((((........))))))..............------ ( -42.70, z-score = -2.80, R) >droAna3.scaffold_12916 378187 110 - 16180835 UAAAAAUGCGAGUUGCGUUGCCUAAUUGCA-GGCGACUGCCUCCAAGGCGGCUUACGUGGCGUAUACGCGACGUGGGGGGCUUU-UCAUUUUCCUUAGCUCGGUUUUUGUCC (((((((.(((((((.(((((((......)-)))))).(((((((((....)))((((.(((....))).)))).))))))...-..........))))))))))))))... ( -42.00, z-score = -2.13, R) >droYak2.chr2L 1757457 97 + 22324452 UCAAAAUGUGACUUGGGUAGCCUAAUUGCG-CACGACUGCCUGGAAGUCGGCUCACGUGGCGUAUACGCAACGUCUGAUAACUU-U--UCUCGCUUUACCC----------- ..............(((((((......(.(-(.(((((.......))))))).)((((.(((....))).))))..........-.--....)))..))))----------- ( -23.90, z-score = 0.01, R) >droSec1.super_14 1716401 97 + 2068291 UAAAAAUGUGACUUGCGUGGCCUAAUUGCG-AACGACUGCCUGGAAGGCGGCUCAUGUGGCGUAUACGCUACGUCUGAUAACUU-U--UCUCGCUUUGCCC----------- .....((((.....))))(((......(((-(..(((((((.....))))).))((((((((....))))))))..........-.--..))))...))).----------- ( -28.40, z-score = -1.54, R) >droSim1.chr2L 20252045 97 - 22036055 UAAAAAUGUGACUUGCGUGGCCUAAUUGCG-AACGACUGCCUGGAAGGCGGCUCACCUGGCGUAUACGCAACGUCUGAUAACUU-U--UCUCGCUUUGCCC----------- .....((((.....))))(((......(((-(..(((((((.....))))).))....(((((.......))))).........-.--..))))...))).----------- ( -22.70, z-score = 0.30, R) >consensus UAAAAAUGUGACUUGCGUUGCCUAAUUGCA_GACGACUGCCUGGAAGGCGGCUCACGUGGCGUAUACGCAACGUCUGAUAACUU_U__UCUCGCUUUACCC___________ ..............(((((((.((..(((.....(((((((.....))))).)).....)))..)).)))))))...................................... (-14.72 = -14.74 + 0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:13 2011