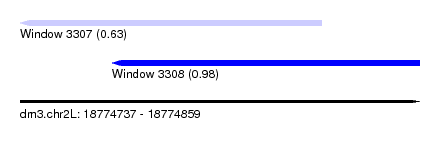
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,774,737 – 18,774,859 |
| Length | 122 |
| Max. P | 0.984817 |

| Location | 18,774,737 – 18,774,829 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 77.99 |
| Shannon entropy | 0.41767 |
| G+C content | 0.56468 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -15.88 |
| Energy contribution | -17.93 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
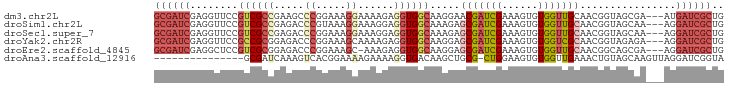
>dm3.chr2L 18774737 92 - 23011544 GCGAUCGAGGUUCCGUCGCCGAAGCCCGGAAAGGAAAAGAGGUGGCAAGGAACGAUCGAAAGUGUGGUUGCAACGGUAGCGA---AUGAUCGCUG ((((((((.(((((((((((.....((.....))......))))))..)))))..)).........(((((....)))))..---..)))))).. ( -32.00, z-score = -1.81, R) >droSim1.chr2L 18460262 92 - 22036055 GCGAUCGAGGUUCCGUCGCCGAGACCCGUAAAGGAAAGGAGGUGGCAAAGAGCGAUCGAAAGUGUGGUUGCAACGGUAGCAA---AGGAUCGCUG ((((((...(((((((((((.....((.....))......)))))).....(((((((......)))))))...)).)))..---..)))))).. ( -32.00, z-score = -2.10, R) >droSec1.super_7 2395335 92 - 3727775 GCGAUCGAGGUUCCGUCGCCGAGACCCGGAAAGGAAAGGAGGUGGCAAAGAGCGAUCGAAAGUGUGGUUGCAACGGUAGCAA---AGGAUCGCUG ((((((...(((((((((((.....((.....))......)))))).....(((((((......)))))))...)).)))..---..)))))).. ( -32.00, z-score = -1.88, R) >droYak2.chr2R 5288547 92 - 21139217 GCGAUCGAGGUUCCGCCGCGGAGACCCGGAAAGCAAAAGAGGUGGCAAGGAGCGAUCGAAAGUGUGGUCGCAACGGUAGAGA---AGGAUCGCUG ((((((....(((..(((..(.((((......(((.....(((.((.....)).))).....))))))).)..)))..))).---..)))))).. ( -31.80, z-score = -1.46, R) >droEre2.scaffold_4845 17087443 91 + 22589142 GCGAUCGAGGCUCCGUCGCGGAGACCCGGAAAGC-AAAGAGGUGGCAAGGAGCGAUCGAAAGUGUGGUUGCAACGGCAGCGA---AGGAUCGCUG ((((((((.(((((.((.(((....)))))..((-.........))..)))))..)).........(((((....)))))..---..)))))).. ( -31.70, z-score = -0.60, R) >droAna3.scaffold_12916 8243949 79 - 16180835 ---------------GCGAUCAAAGUCACGGAAAAGAAAAGGUGACAAGCUGCG-CUGGAAGUGUGGUUGAAACUGUAGCAAGUUAGGAUCGGUA ---------------.(((((...(((((............))))).(((..((-(.....)))..)))..((((......))))..)))))... ( -21.90, z-score = -2.32, R) >consensus GCGAUCGAGGUUCCGUCGCCGAGACCCGGAAAGGAAAAGAGGUGGCAAGGAGCGAUCGAAAGUGUGGUUGCAACGGUAGCAA___AGGAUCGCUG ((((((........((((((.....((.....))......)))))).....(((((((......)))))))................)))))).. (-15.88 = -17.93 + 2.06)
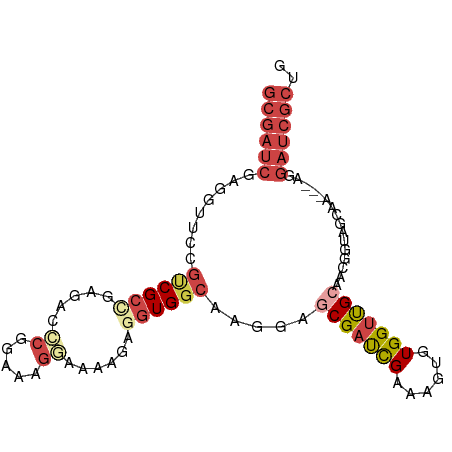
| Location | 18,774,765 – 18,774,859 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 72.19 |
| Shannon entropy | 0.58310 |
| G+C content | 0.59832 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.66 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18774765 94 - 23011544 GUGGCACCAGUGCAGCACGGGGGUGGCACUGCGAUCGAGGUUCCGUCGCCGAAGCCCGGAAAGGAAAAGAGGUGGCAAGGAACGAUCGAAAGUG (((.((((.(((...)))...)))).))).((..((((.(((((((((((.....((.....))......))))))..)))))..))))..)). ( -35.90, z-score = -1.78, R) >droSim1.chr2L 18460290 94 - 22036055 GUGCCACCAGUGCAGCACGGGGGUGGCAACGCGAUCGAGGUUCCGUCGCCGAGACCCGUAAAGGAAAGGAGGUGGCAAAGAGCGAUCGAAAGUG .(((((((.(((...)))...))))))).(((..((((.((((.((((((.....((.....))......))))))...))))..))))..))) ( -35.90, z-score = -1.98, R) >droSec1.super_7 2395363 94 - 3727775 GUGCCACCAGUGCAGCACGGGGGUGGCAACGCGAUCGAGGUUCCGUCGCCGAGACCCGGAAAGGAAAGGAGGUGGCAAAGAGCGAUCGAAAGUG .(((((((.(((...)))...))))))).(((..((((.((((.((((((.....((.....))......))))))...))))..))))..))) ( -35.90, z-score = -1.75, R) >droYak2.chr2R 5288575 94 - 21139217 GUGCCACCAGUGCAGCACGGGGGUGGCAACGCGAUCGAGGUUCCGCCGCGGAGACCCGGAAAGCAAAAGAGGUGGCAAGGAGCGAUCGAAAGUG .(((((((.(((...)))...))))))).(((..((((.((((((((((((....))(.....).......)))))..)))))..))))..))) ( -36.70, z-score = -1.68, R) >droEre2.scaffold_4845 17087471 93 + 22589142 GUGCCACCAGUGCAGCACGGGGGUGGCAACGCGAUCGAGGCUCCGUCGCGGAGACCCGGAAAGC-AAAGAGGUGGCAAGGAGCGAUCGAAAGUG .(((((((.(((...)))...))))))).(((..((((.(((((.((.(((....)))))..((-.........))..)))))..))))..))) ( -36.80, z-score = -1.60, R) >droAna3.scaffold_12916 8243980 76 - 16180835 --GCCACUAGUGCAGCACGGGGGUGGCAACGCGAUCAAAGU-------------CACGGAAAAGAAA--AGGUGACAAGCUGCGCU-GGAAGUG --....((((((((((..((..(((....)))..))...((-------------(((..........--..)))))..))))))))-))..... ( -30.90, z-score = -3.84, R) >droGri2.scaffold_15252 11215937 88 + 17193109 CGAUCCUUAAUGCAGCUAAACAGGGUUGA----AACUGGUUGCCGCCAGUCAAAUGCUAAGCAUCGUUGAGUUG--AAGGUGGAAUUGGAGGUG (((((((..............))))))).----..(..(((.((((((.((((((((...))))..)))).)..--..))))))))..)..... ( -20.64, z-score = 0.34, R) >consensus GUGCCACCAGUGCAGCACGGGGGUGGCAACGCGAUCGAGGUUCCGUCGCCGAGACCCGGAAAGGAAAAGAGGUGGCAAGGAGCGAUCGAAAGUG ..((((((.(((...)))...))))))..(((..((((.((((.(((((......................)))))...))))..))))..))) (-18.29 = -18.66 + 0.38)

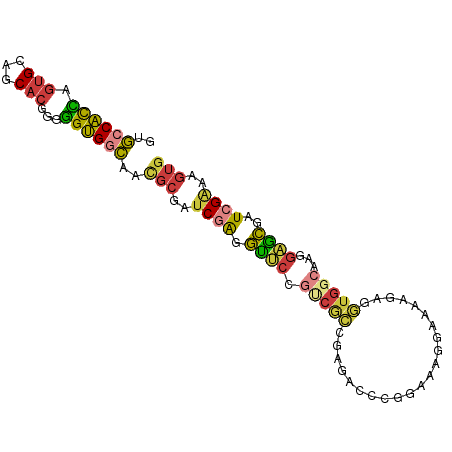
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:35 2011