
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,744,268 – 18,744,388 |
| Length | 120 |
| Max. P | 0.731342 |

| Location | 18,744,268 – 18,744,388 |
|---|---|
| Length | 120 |
| Sequences | 10 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 72.09 |
| Shannon entropy | 0.59327 |
| G+C content | 0.43029 |
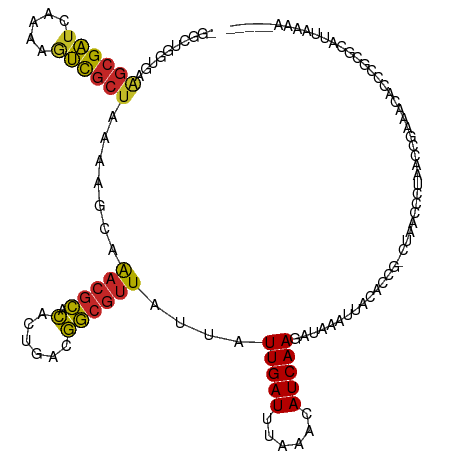
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -11.47 |
| Energy contribution | -11.29 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
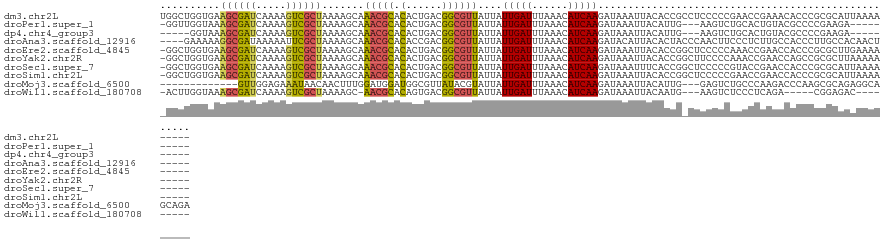
>dm3.chr2L 18744268 120 - 23011544 UGGCUGGUGAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACACCGCCUCCCCCGAACCGAAACACCCGCGCAUUAAAA----- ..((.((((.((((((.....))))))....((....))........(((((((((.(((((......)))))))))).......))))...............)))).)).........----- ( -25.01, z-score = -1.12, R) >droPer1.super_1 6221117 111 - 10282868 -GGUUGGUAAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACAUUG---AAGUCUGCACUGUACGCCCCGAAGA---------- -.((..((..((((((.....))))))....((....))..))..))(((((((((.(((((......))))))))))..((((.((---.......)).)))))))).......---------- ( -24.90, z-score = -1.00, R) >dp4.chr4_group3 4729398 107 - 11692001 -----GGUAAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACAUUG---AAGUCUGCACUGUACGCCCCGAAGA---------- -----((...((((((.....))))))....((....))........(((((((((.(((((......))))))))))..((((.((---.......)).)))))))))).....---------- ( -20.30, z-score = -0.26, R) >droAna3.scaffold_12916 8216274 116 - 16180835 ----GAAAAAGGCGAUAAAAAUUCGCUAAAAGCAAACGCACACCGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUACAUUACACUACCCAACUUCCCUCUUGCCACCCUUGCCACAACU----- ----......(((((.......)))))....((((((((.(......))))))....(((((......)))))............................)))................----- ( -15.20, z-score = -1.13, R) >droEre2.scaffold_4845 17056889 119 + 22589142 -GGCUGGUGAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACACCGGCUCCCCCAAACCGAACCACCCGCGCUUGAAAA----- -((((((((.((((((.....)))))).......(((((.(......))))))....(((((......))))).........))))))))....(((.(((.......)).).)))....----- ( -27.20, z-score = -1.46, R) >droYak2.chr2R 5257873 119 - 21139217 -GGCUGGUGAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACACCGGCUUCCCCAAACCGAACCAGCCGCGCUUAAAAA----- -(((((((..((((((.....))))))..((((.(((((.(......))))))....(((((......)))))..............))))...........)))))))...........----- ( -31.00, z-score = -2.44, R) >droSec1.super_7 2365509 119 - 3727775 -GGCUGGUGAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUUCACCGGCUCCCCCGUACCGAACCACCCGCGCAUUAAAA----- -(((((((((((((((.....)))))).......(((((.(......))))))....(((((......)))))........)))))))))..............................----- ( -27.70, z-score = -1.25, R) >droSim1.chr2L 18431894 119 - 22036055 -GGCUGGUGAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACACCGGCUCCCCCGAACCGAACCACCCGCGCAUUAAAA----- -((((((((.((((((.....)))))).......(((((.(......))))))....(((((......))))).........))))))))..............................----- ( -26.40, z-score = -1.20, R) >droMoj3.scaffold_6500 5697488 109 + 32352404 -------------GUUGGAGAAAUAACAACUUUGGAUGGAUGGCGUUAUACGUAUUAUUGAUUUAAACAUCAAGAUAAAUUACAUUG---GAGUCUGCCCAAGACCCAAGCGCAGAGGCAGCAGA -------------((((.........))))(((((((.(((((((.....)))..)))).))))))).................(((---(.((((.....))))))))((((....)).))... ( -21.40, z-score = -0.09, R) >droWil1.scaffold_180708 1418786 106 + 12563649 -ACUUGGUAAAGCGAUCAAAAGUCGCUAAAAGC-AACGCACAGUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACAAUG---AAGUCUCCCUCAGA-----CGGAGAC--------- -.((((((..((((((.....))))))......-(((((.(......))))))...............)))))).............---..((((((......-----.))))))--------- ( -24.90, z-score = -2.10, R) >consensus _GGCUGGUGAAGCGAUCAAAAGUCGCUAAAAGCAAACGCACACUGACGGCGUUAUUAUUGAUUUAAACAUCAAGAUAAAUUACACCG_CUAACCCUAACCGAAACACCCGCGCAUUAAAA_____ ..........((((((.....)))))).......(((((.(......))))))....(((((......))))).................................................... (-11.47 = -11.29 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:32 2011