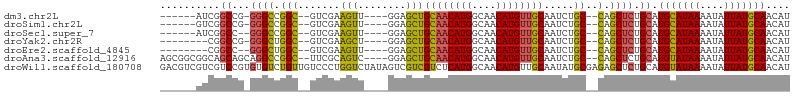
| Sequence ID | dm3.chr2L |
|---|---|
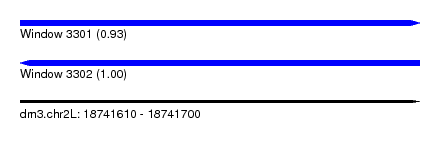
| Location | 18,741,610 – 18,741,700 |
| Length | 90 |
| Max. P | 0.998632 |

| Location | 18,741,610 – 18,741,700 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.31184 |
| G+C content | 0.50719 |
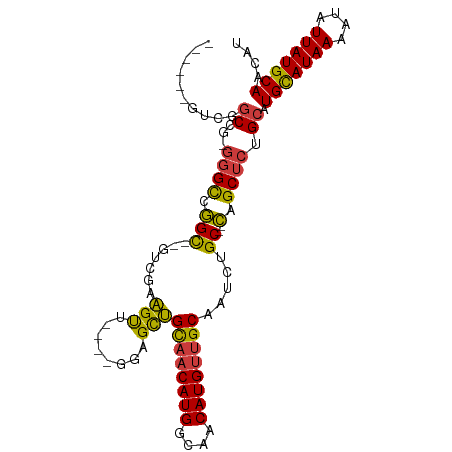
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -21.13 |
| Energy contribution | -21.99 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
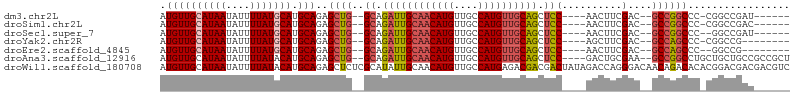
>dm3.chr2L 18741610 90 + 23011544 AUGUUGCAUAAUAUUUUAUGCAUGCAGAGCUG--GCAGAUUGCAACAUGUUGCCAUGUUGCAGCUCC----AACUUCGAC--GCCGGCCC-CGGCCGAU------ .((((((((((....))))))).)))(((.((--(.((.((((((((((....))))))))))))))----).)))....--..((((..-..))))..------ ( -35.20, z-score = -2.97, R) >droSim1.chr2L 18429080 90 + 22036055 AUGUUGCAUAAUAUUUUAUGCAUGCAGAGCUG--GCAGAUUGCAACAUGUUGCCAUGUUGCAGCUCC----AACUUCGAC--GCCGGCCC-CGGCCGAC------ .((((((((((....))))))).)))(((.((--(.((.((((((((((....))))))))))))))----).)))....--..((((..-..))))..------ ( -35.20, z-score = -3.17, R) >droSec1.super_7 2362840 89 + 3727775 AUGUUGCAUAAUAUUUUAUGCAUGCAGAGCUG--GCAGAUUGCAACAUGUUGCCAUGUUGCAGCUCC----AACUUCGAC--GCCGGCCC--GGCCGAU------ .((((((((((....))))))).)))(((.((--(.((.((((((((((....))))))))))))))----).)))....--..((((..--.))))..------ ( -32.20, z-score = -1.98, R) >droYak2.chr2R 5255205 88 + 21139217 AUGUUGCAUAAUAUUUUAUGCAUGCAGAGCUG--GCAGAUUGCAACAUGUUGCCAUGUUGCAGCUCC----AGCUUCGAC--GCCAGCCC-CGGCCG-------- .((((((((((....))))))).)))((((((--(.((.((((((((((....))))))))))))))----)))))....--(((.....-.)))..-------- ( -37.90, z-score = -4.15, R) >droEre2.scaffold_4845 17054120 87 - 22589142 AUGUUGCAUAAUAUUUUAUGCAUGCAGAGCUG--GCAGAUUGCAACAUGUUGCCAUGUUGCAGCUCC----AACUUCGAC--GCCAGCCC--GGCCG-------- ....(((((((....))))))).((.(.((((--((.((((((((((((....)))))))))).))(----......)..--)))))).)--.))..-------- ( -32.40, z-score = -2.71, R) >droAna3.scaffold_12916 8214244 97 + 16180835 AUGUUGCAUAAUAUUUUAUACAUGCAGAGCUG--GCAGAUUGCAACAUGUUGCCAUGUUGCAGCUCC----GACUGCGAA--GCCGGCCUGCUGCUGCCGCCGCU ..((.((((((....)))).((.((((.((((--((....(((((((((....)))))))))((...----....))...--)))))))))))).....)).)). ( -35.30, z-score = -1.24, R) >droWil1.scaffold_180708 1415789 105 - 12563649 AUGUUGCAUAAUAUUUUAUACAUGCAGAGCUCUCGCAUAUUGCAACAUGUUGCCAUGAGACGACGACUAUAGACCAGGGACAACAGACACACGGACGACGACGUC .((((((((((....))))..((((((....)).))))...))))))(((((((.((.................)).)).)))))........((((....)))) ( -20.23, z-score = -0.12, R) >consensus AUGUUGCAUAAUAUUUUAUGCAUGCAGAGCUG__GCAGAUUGCAACAUGUUGCCAUGUUGCAGCUCC____AACUUCGAC__GCCGGCCC_CGGCCGAC______ .((((((((((....))))))).)))..((....)).((((((((((((....)))))))))).))..................((((.....))))........ (-21.13 = -21.99 + 0.86)
| Location | 18,741,610 – 18,741,700 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.31184 |
| G+C content | 0.50719 |
| Mean single sequence MFE | -39.16 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.64 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.998632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
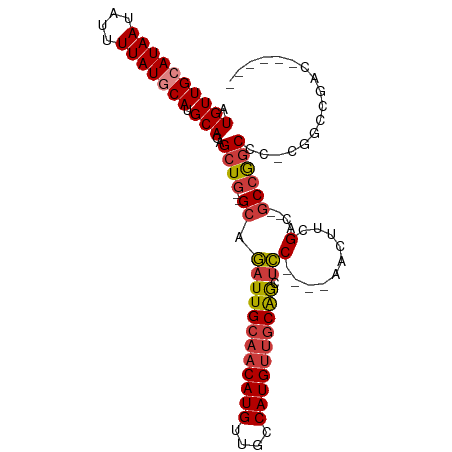
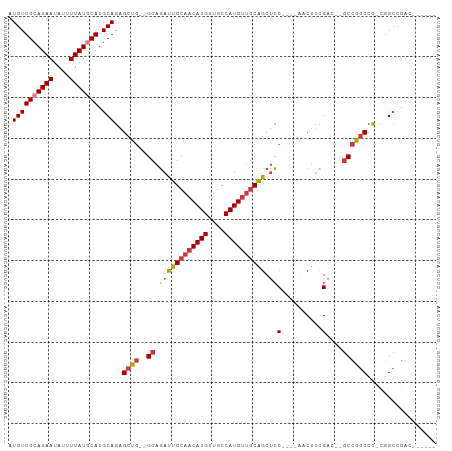
>dm3.chr2L 18741610 90 - 23011544 ------AUCGGCCG-GGGCCGGC--GUCGAAGUU----GGAGCUGCAACAUGGCAACAUGUUGCAAUCUGC--CAGCUCUGCAUGCAUAAAAUAUUAUGCAACAU ------.(((((..-..))))).--((.((.(((----((((.(((((((((....)))))))))..)).)--)))))).)).(((((((....))))))).... ( -40.00, z-score = -3.86, R) >droSim1.chr2L 18429080 90 - 22036055 ------GUCGGCCG-GGGCCGGC--GUCGAAGUU----GGAGCUGCAACAUGGCAACAUGUUGCAAUCUGC--CAGCUCUGCAUGCAUAAAAUAUUAUGCAACAU ------((((((..-..))))))--((.((.(((----((((.(((((((((....)))))))))..)).)--)))))).)).(((((((....))))))).... ( -43.90, z-score = -4.99, R) >droSec1.super_7 2362840 89 - 3727775 ------AUCGGCC--GGGCCGGC--GUCGAAGUU----GGAGCUGCAACAUGGCAACAUGUUGCAAUCUGC--CAGCUCUGCAUGCAUAAAAUAUUAUGCAACAU ------.(((((.--..))))).--((.((.(((----((((.(((((((((....)))))))))..)).)--)))))).)).(((((((....))))))).... ( -37.00, z-score = -3.15, R) >droYak2.chr2R 5255205 88 - 21139217 --------CGGCCG-GGGCUGGC--GUCGAAGCU----GGAGCUGCAACAUGGCAACAUGUUGCAAUCUGC--CAGCUCUGCAUGCAUAAAAUAUUAUGCAACAU --------..((.(-((((((((--.....(((.----...)))((((((((....)))))))).....))--))))))))).(((((((....))))))).... ( -43.00, z-score = -4.94, R) >droEre2.scaffold_4845 17054120 87 + 22589142 --------CGGCC--GGGCUGGC--GUCGAAGUU----GGAGCUGCAACAUGGCAACAUGUUGCAAUCUGC--CAGCUCUGCAUGCAUAAAAUAUUAUGCAACAU --------..((.--((((((((--.........----(((..(((((((((....))))))))).)))))--)))))).)).(((((((....))))))).... ( -41.20, z-score = -4.75, R) >droAna3.scaffold_12916 8214244 97 - 16180835 AGCGGCGGCAGCAGCAGGCCGGC--UUCGCAGUC----GGAGCUGCAACAUGGCAACAUGUUGCAAUCUGC--CAGCUCUGCAUGUAUAAAAUAUUAUGCAACAU ....((((.(((.((((((((((--(....))))----))...(((((((((....))))))))).)))))--..))))))).(((((((....))))))).... ( -42.90, z-score = -3.38, R) >droWil1.scaffold_180708 1415789 105 + 12563649 GACGUCGUCGUCCGUGUGUCUGUUGUCCCUGGUCUAUAGUCGUCGUCUCAUGGCAACAUGUUGCAAUAUGCGAGAGCUCUGCAUGUAUAAAAUAUUAUGCAACAU ((((.((.(..(((.(.(.(....).)).)))......).)).)))).((((....)))).((((....((....))..))))(((((((....))))))).... ( -26.10, z-score = -0.51, R) >consensus ______GUCGGCCG_GGGCCGGC__GUCGAAGUU____GGAGCUGCAACAUGGCAACAUGUUGCAAUCUGC__CAGCUCUGCAUGCAUAAAAUAUUAUGCAACAU ........((((.....)))).........((((....(((..(((((((((....))))))))).))).....)))).....(((((((....))))))).... (-27.33 = -27.64 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:30 2011