| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,774,579 – 1,774,723 |
| Length | 144 |
| Max. P | 0.629777 |

| Location | 1,774,579 – 1,774,683 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Shannon entropy | 0.44768 |
| G+C content | 0.44216 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -14.12 |
| Energy contribution | -13.45 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
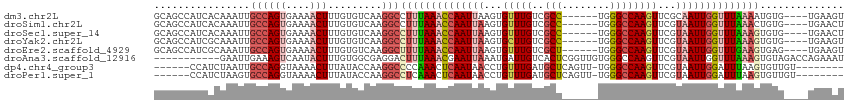
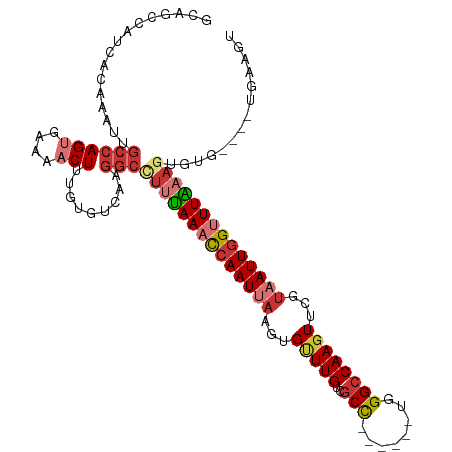
>dm3.chr2L 1774579 104 + 23011544 -UAAACUUGAAACGGGGUACCCAUAUGCCACCCUCUUCUCACUUCACACAUUUUAAACCAAUUGCGAACUUGGCCCAG-----GCGACAAACACUUAAUUGGUUUAAAGG -.......(((..((((((......)))).))...)))............((((((((((((((.(...(((((....-----))..)))...).)))))))))))))). ( -20.50, z-score = -0.90, R) >droSim1.chr2L 1762116 104 + 22036055 -UCAACUUGAAACGGGGUACCCAUAUGCCACCCUCUUCUCAGUUCACACAGUUUAAACCAAUUACGAACUUGGCCCAG-----GCGACAAACACUUAAUUGGUUUAAAGG -..((((.(((..((((((......)))).))...)))..)))).......(((((((((((((.(...(((((....-----))..)))...).))))))))))))).. ( -21.50, z-score = -0.65, R) >droSec1.super_14 1709030 101 + 2068291 ----ACUUGAAACGGGGUACCCAUACGCCACCCUCUUCUCAGUUCACACACUUUAAACCAAUUACGAACUUGGCCCAG-----GCGACAAACACUUAAUUGGUUUAAAGG ----..((((((.(((((...........))))).)).))))........((((((((((((((.(...(((((....-----))..)))...).)))))))))))))). ( -23.90, z-score = -2.13, R) >droYak2.chr2L 1748030 103 + 22324452 -UUAACUUGAAACGGGGUAACCGUAUGCCACCCU-UUCACACUUCACACACUUUAAACCAAUUACGAACUUGGCCCAG-----GCGACAAGCACUUAAUUGGUUUAAAGG -......(((((.((((((......)))).)).)-))))...........((((((((((((((.(..((((((....-----))..))))..).)))))))))))))). ( -31.20, z-score = -4.52, R) >droEre2.scaffold_4929 1820934 104 + 26641161 -UCAACUUGAAAUGGGGUAACCAUAUGCCACCCUUUUCCCACUUCACUCACUUCAAACCAAUUACGAACUUGGCCCAA-----GCGACAAACACUUAAUUGGUUUAAAAG -......((((.(((((...................))))).))))........((((((((((.(...(((((....-----))..)))...).))))))))))..... ( -17.31, z-score = -0.98, R) >dp4.chr4_group3 3155957 105 - 11692001 GUGUGUUUAACAUGGAGCUCUCCCA-GUCAUCCCCCUCUGAC----AACACUUAAAUCCAAUUACGAACUUGGCCCAAACUGAGCAUCAAACAGGUUAUUGAGUUUGGGG ..(((((......(((....)))..-((((........))))----)))))......((((........))))((((((((.((.(((.....)))..)).)))))))). ( -22.90, z-score = 0.19, R) >droPer1.super_1 296857 105 - 10282868 GUGUGUUCAACAUGGAGCUCUCCCA-GUCAUCCCCCUCUGAC----AACACUUAAAUCCAAUUACGAACUUGGCCCAAACUGAGCAUCAAACAGGUUAUUGAGUUUGAGG ((((.....))))(((....)))..-((((........))))----....((((((..((((....((((((...................))))))))))..)))))). ( -18.31, z-score = 1.06, R) >consensus _UCAACUUGAAACGGGGUACCCAUAUGCCACCCUCUUCUCACUUCACACACUUUAAACCAAUUACGAACUUGGCCCAG_____GCGACAAACACUUAAUUGGUUUAAAGG .............(((..............))).................((((((((((((((.....(((((.........))..))).....)))))))))))))). (-14.12 = -13.45 + -0.67)


| Location | 1,774,579 – 1,774,683 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.77 |
| Shannon entropy | 0.44768 |
| G+C content | 0.44216 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -16.35 |
| Energy contribution | -16.69 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.574767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1774579 104 - 23011544 CCUUUAAACCAAUUAAGUGUUUGUCGC-----CUGGGCCAAGUUCGCAAUUGGUUUAAAAUGUGUGAAGUGAGAAGAGGGUGGCAUAUGGGUACCCCGUUUCAAGUUUA- ..((((((((((((..(..((((..((-----....))))))..)..))))))))))))......((..(((((.(.(((((.(.....).)))))).)))))..))..- ( -28.00, z-score = -1.12, R) >droSim1.chr2L 1762116 104 - 22036055 CCUUUAAACCAAUUAAGUGUUUGUCGC-----CUGGGCCAAGUUCGUAAUUGGUUUAAACUGUGUGAACUGAGAAGAGGGUGGCAUAUGGGUACCCCGUUUCAAGUUGA- ..(((((((((((((.(..((((..((-----....))))))..).))))))))))))).......((((((((.(.(((((.(.....).)))))).)))).))))..- ( -30.10, z-score = -1.43, R) >droSec1.super_14 1709030 101 - 2068291 CCUUUAAACCAAUUAAGUGUUUGUCGC-----CUGGGCCAAGUUCGUAAUUGGUUUAAAGUGUGUGAACUGAGAAGAGGGUGGCGUAUGGGUACCCCGUUUCAAGU---- .((((((((((((((.(..((((..((-----....))))))..).)))))))))))))).........(((((.(.(((((.(.....).)))))).)))))...---- ( -31.60, z-score = -2.50, R) >droYak2.chr2L 1748030 103 - 22324452 CCUUUAAACCAAUUAAGUGCUUGUCGC-----CUGGGCCAAGUUCGUAAUUGGUUUAAAGUGUGUGAAGUGUGAA-AGGGUGGCAUACGGUUACCCCGUUUCAAGUUAA- .((((((((((((((...(((((..((-----....)))))))...))))))))))))))....(((..((.((.-.(((((((.....)))))))..)).))..))).- ( -36.30, z-score = -4.02, R) >droEre2.scaffold_4929 1820934 104 - 26641161 CUUUUAAACCAAUUAAGUGUUUGUCGC-----UUGGGCCAAGUUCGUAAUUGGUUUGAAGUGAGUGAAGUGGGAAAAGGGUGGCAUAUGGUUACCCCAUUUCAAGUUGA- .((((((((((((((.(..((((..((-----....))))))..).))))))))))))))....(((((((((......(((((.....))))))))))))))......- ( -30.60, z-score = -2.34, R) >dp4.chr4_group3 3155957 105 + 11692001 CCCCAAACUCAAUAACCUGUUUGAUGCUCAGUUUGGGCCAAGUUCGUAAUUGGAUUUAAGUGUU----GUCAGAGGGGGAUGAC-UGGGAGAGCUCCAUGUUAAACACAC ((((...(((.(((((((.......((((.....)))).(((((((....))))))).)).)))----))..))))))).((((-((((.....)))).))))....... ( -24.70, z-score = 0.91, R) >droPer1.super_1 296857 105 + 10282868 CCUCAAACUCAAUAACCUGUUUGAUGCUCAGUUUGGGCCAAGUUCGUAAUUGGAUUUAAGUGUU----GUCAGAGGGGGAUGAC-UGGGAGAGCUCCAUGUUGAACACAC ........((((((.(((.((((((((((.....)))).(((((((....))))))).......----)))))).)))......-..(((....))).))))))...... ( -25.50, z-score = 0.71, R) >consensus CCUUUAAACCAAUUAAGUGUUUGUCGC_____CUGGGCCAAGUUCGUAAUUGGUUUAAAGUGUGUGAAGUGAGAAGAGGGUGGCAUAUGGGUACCCCGUUUCAAGUUCA_ .((((((((((((((.(..((((.(.(.......).).))))..).))))))))))))))............((((.(((.(...........)))).))))........ (-16.35 = -16.69 + 0.33)


| Location | 1,774,618 – 1,774,723 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Shannon entropy | 0.47561 |
| G+C content | 0.41116 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -13.99 |
| Energy contribution | -15.18 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1774618 105 - 23011544 GCAGCCAUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCC------UGGGCCAAGUUCGCAAUUGGUUUAAAAUGUG----UGAAGU .......(((((.((((((((((..(((((.(.(((.((((...((((((......).)))))..)))------).)))))))))..).))))))....))).))----)))... ( -26.90, z-score = -0.98, R) >droSim1.chr2L 1762155 105 - 22036055 GCAGCCAUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCC------UGGGCCAAGUUCGUAAUUGGUUUAAACUGUG----UGAACU ((((((..(((((......(((....))))))))....))).(((((((((((((.(..((((..((.------...))))))..).)))))))))))))...))----)..... ( -26.30, z-score = -0.86, R) >droSec1.super_14 1709066 105 - 2068291 GCAGCCAUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCC------UGGGCCAAGUUCGUAAUUGGUUUAAAGUGUG----UGAACU ((((((..(((((......(((....))))))))....)))((((((((((((((.(..((((..((.------...))))))..).))))))))))))))))).----...... ( -29.20, z-score = -1.84, R) >droYak2.chr2L 1748068 105 - 22324452 GCAGCCAUCGCAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGCUUGUCGCC------UGGGCCAAGUUCGUAAUUGGUUUAAAGUGUG----UGAAGU ((((((..(((((......(((....))))))))....)))((((((((((((((...(((((..((.------...)))))))...))))))))))))))))).----...... ( -32.80, z-score = -2.75, R) >droEre2.scaffold_4929 1820973 105 - 26641161 GCAGCCAUCGCAAAUUGCCAGUGAAAACUUUGUGUCAAGGCUUUUAAACCAAUUAAGUGUUUGUCGCU------UGGGCCAAGUUCGUAAUUGGUUUGAAGUGAG----UGAAGU ..((((..(((((......(((....))))))))....))))(((((((((((((.(..((((..((.------...))))))..).))))))))))))).....----...... ( -25.60, z-score = -0.50, R) >droAna3.scaffold_12916 369907 104 + 16180835 -----------GAAUUGAAAGUCAAUACUUUGUGGCGAGGACUUUAAACGAAUUAAAUGAUUGUCACUCGGUUGUGGGCCAAGUUCGUAAUUGGUUUAAAGUGUAGACCAGAAAU -----------..((((.....))))..(((.((((..(.(((((((((.(((((.....(((((((......))))..))).....))))).))))))))).).).))).))). ( -24.10, z-score = -1.25, R) >dp4.chr4_group3 3155996 100 + 11692001 ------CCAUCUAAUUGCCAGGUAAAACUUUAUACCAAGGCCCCAAACUCAAUAACCUGUUUGAUGCUCAGUU-UGGGCCAAGUUCGUAAUUGGAUUUAAGUGUUGU-------- ------..((((((((((..((((........))))..(((((.((((((((........))))......)))-))))))......))))))))))...........-------- ( -26.30, z-score = -1.94, R) >droPer1.super_1 296896 100 + 10282868 ------CCAUCUAAGUGCCAGGUAAAACUUUAUACCAAGGCCUCAAACUCAAUAACCUGUUUGAUGCUCAGUU-UGGGCCAAGUUCGUAAUUGGAUUUAAGUGUUGU-------- ------.((..(((((.(((((((........))))..((((.(((((((((........))))......)))-))))))...........))))))))..))....-------- ( -23.80, z-score = -1.10, R) >consensus GCAGCCAUCACAAAUUGCCAGUGAAAACUUUGUGUCAAGGCCUUUAAACCAAUUAAGUGUUUGUCGCC______UGGGCCAAGUUCGUAAUUGGUUUAAAGUGUG____UGAAGU ................((((((....))).........)))((((((((((((((...(((((..(((........))))))))...)))))))))))))).............. (-13.99 = -15.18 + 1.19)
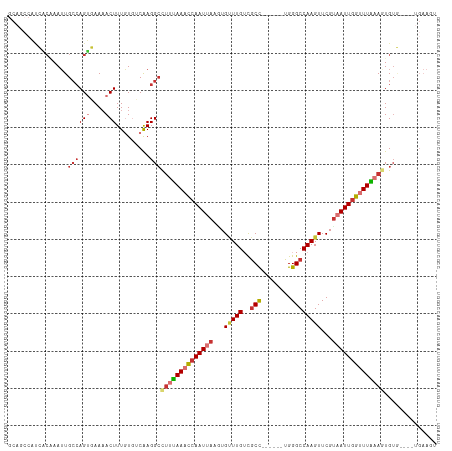
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:11 2011