
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,663,698 – 18,663,818 |
| Length | 120 |
| Max. P | 0.516072 |

| Location | 18,663,698 – 18,663,818 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 137 |
| Reading direction | reverse |
| Mean pairwise identity | 74.18 |
| Shannon entropy | 0.55275 |
| G+C content | 0.42964 |
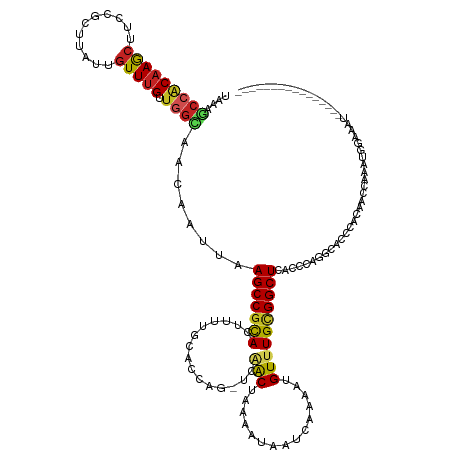
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.85 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
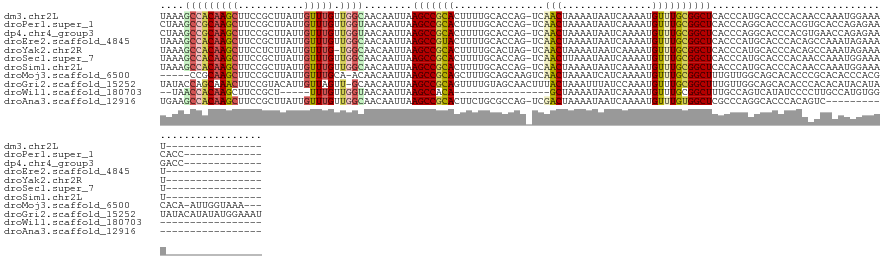
>dm3.chr2L 18663698 120 - 23011544 UAAAGCCACAAGCUUCCGCUUAUUGUUUGUUGGCAACAAUUAAGCCGCACUUUUGCACCAG-UCAACUAAAAUAAUCAAAAUGUUUGCGGCUCACCCAUGCACCCACAACCAAAUGGAAAU---------------- ...((((.(((((....((((((((((.(....)))))).))))).(((....))).....-....................))))).))))...((((..............))))....---------------- ( -23.44, z-score = -0.44, R) >droPer1.super_1 6480096 123 + 10282868 CUAAGCCGCAAGCUUCCGCUUAUUGUUUGUUGGUAACAAUUAAGCCGCACUUUUGCACCAG-UCAACUAAAAUAAUCAAAAUGUUUGCGGCUCACCCAGGCACCCACGUGCACCAGAGAACACC------------- ...((((((((((....((((((((((.......))))).))))).(((....))).....-....................)))))))))).......((((....)))).............------------- ( -31.10, z-score = -1.72, R) >dp4.chr4_group3 9362056 123 + 11692001 CUAAGCCGCAAGCUUCCGCUUAUUGUUUGUUGGUAACAAUUAAGCCGCACUUUUGCACCAG-UCAACUAAAAUAAUCAAAAUGUUUGCGGCUCACCCAGGCACCCACGUGAACCAGAGAAGACC------------- ...((((((((((....((((((((((.......))))).))))).(((....))).....-....................))))))))))......(((((....)))..))..........------------- ( -28.40, z-score = -0.91, R) >droEre2.scaffold_4845 16976426 120 + 22589142 UAAAGCCACAAGCUUCCGCUUAUUGUUUGUUGGCAACAAUUAAGCCGUACUUUUGCACCAG-UCAACUAAAAUAAUCAAAAUGUUUGCGGCUCACCCAUGCACCCACAGCCAAAUAGAAAU---------------- ...((((.(((((....((((((((((.(....)))))).))))).(((....))).....-....................))))).)))).............................---------------- ( -19.00, z-score = 0.68, R) >droYak2.chr2R 5176992 119 - 21139217 UAAAGCCACAAGCUUCCUCUUAUUGUUUG-UGGCAACAAUUAAGCCGCACUUUUGCACUAG-UCAACUAAAAUAAUCAAAAUGUUUGCGGCUCACCCAUGCACCCACAGCCAAAUAGAAAU---------------- ....(((((((((...........)))))-))))........(((((((.(((((...(((-....))).......)))))....))))))).............................---------------- ( -25.60, z-score = -1.91, R) >droSec1.super_7 2285298 120 - 3727775 UAAAGCCACAAGCUUCCGCUUAUUGUUUGUUGGCAACAAUUAAGCCGCACUUUUGCACCAG-UCAACUUAAAUAAUCAAAAUGUUUGCGGCUCACCCAUGCACCCACAACCAAAUGGAAAU---------------- ...((((.(((((....((((((((((.(....)))))).))))).(((....))).....-....................))))).))))...((((..............))))....---------------- ( -23.44, z-score = -0.39, R) >droSim1.chr2L 18351990 120 - 22036055 UAAAGCCACAAGCUUCCGCUUAUUGUUUGUUGGCAACAAUUAAGCCGCACUUUUGCACCAG-UCAACUAAAAUAAUCAAAAUGUUUGCGGCUCACCCAUGCACCCACAACCAAAUGGAAAU---------------- ...((((.(((((....((((((((((.(....)))))).))))).(((....))).....-....................))))).))))...((((..............))))....---------------- ( -23.44, z-score = -0.44, R) >droMoj3.scaffold_6500 7921464 127 + 32352404 -----CCGCAAGCUUCCGCUUAUUGUUUGCA-ACAACAAUUAAGCCGCAGCUUUGCAGCAAGUCAACUAAAAUCAUCAAAAUGUUUGCGGCUUUGUUGGCAGCACACCCGCACACCCACGCACA-AUUGGUAAA--- -----..((((((....))))...((.(((.-.((((((...((((((((((....))).((....)).................)))))))))))))...))).))..))....(((......-..)))....--- ( -29.40, z-score = 0.42, R) >droGri2.scaffold_15252 6647527 136 - 17193109 UAUACCAGCAAACUUCCGUACAUUGUUAGUU-GCAACAAUUAAGCCGCAGUUUUGUAGCAACUUUACUAAAUUUAUCCAAAUGUUUGCGGCUUUGUUGGCAGCACACCCACACAUACAUAUAUACAUAUAUGGAAAU .............(((((((...(((..(((-((..((((.(((((((((.(((((((........)))........))))...))))))))).)))))))))..........(((....))))))..))))))).. ( -28.50, z-score = -1.09, R) >droWil1.scaffold_180703 1620244 97 - 3946847 --UAACCACAAGCUUCCGCU-----UUUGUUGGUAACAAUUAAGCCACA----------------GCUAAAAUAAUCAAAAUGUUUGCGGCUUUGCCAGUCAUAUCCCCUUGCCAUGUGG----------------- --...(((((((((...(((-----(..((((....)))).))))...)----------------)))....................(((....................))).)))))----------------- ( -19.95, z-score = -0.56, R) >droAna3.scaffold_12916 2554359 110 - 16180835 UGAAGCCACAAGCUUCCGCUUAUUGUUUGUUGGCAACAAUUAAGCCGCACUUCUGCGCCAG-UCGACUAAAAUAAUCAAAAUGUUUGUGGCUCGCCCAGGCACCCACAGUC-------------------------- .((.(((((((((....((((((((((.(....)))))).)))))((((....))))....-....................)))))))))))((....))..........-------------------------- ( -29.10, z-score = -1.26, R) >consensus UAAAGCCACAAGCUUCCGCUUAUUGUUUGUUGGCAACAAUUAAGCCGCACUUUUGCACCAG_UCAACUAAAAUAAUCAAAAUGUUUGCGGCUCACCCAGGCACCCACAACCAAAUGGAAAU________________ ....(((((((((...........)))))).)))........(((((((....................................)))))))............................................. (-14.75 = -14.85 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:20 2011