
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,610,599 – 18,610,689 |
| Length | 90 |
| Max. P | 0.835708 |

| Location | 18,610,599 – 18,610,689 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 63.65 |
| Shannon entropy | 0.71399 |
| G+C content | 0.55115 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -13.17 |
| Energy contribution | -11.97 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18610599 90 - 23011544 ---------------AGCAAAUCCUCUAUCCUAGAGCCUUGCAAACCGAGUGGAUUUGUCAGACAACCACGGCGAAAUUGGCCCUGGUUGCAGGAGG---CGCAGGAU--------- ---------------............(((((.(.((((((((((((....)).)))))..(.((((((.(((.......))).)))))))..))))---).))))))--------- ( -29.90, z-score = -1.21, R) >droGri2.scaffold_15252 6588707 102 - 17193109 ---GCAGAGAGUAGCAGC------AUAAGUUUAGAGCGCUGCAAGCCGAGUGGAUUUCUUCGCCAGCCCCGACGCAAUUGGCCCUGGCUGCAGGAGACCGACGAGCAAGAG------ ---.......(((((.((------...........)))))))..((((..(((.((((((((((((..((((.....))))..))))).).))))))))).)).)).....------ ( -30.30, z-score = 0.12, R) >droMoj3.scaffold_6500 26357999 105 + 32352404 ---------GAAGUAAAAGGCAACAUCAAUCUGGAGCCUUGUAAGCCAAGCGGCUUUAUAAGGCAGCCGCGUCGAAACUGGCCCUGGCUGCAGGAGACGGCAAUAGAGCCGGUC--- ---------.........(((....((......))((((((((((((....))).))))))))).)))........((((((.(((..(((.(....).))).))).)))))).--- ( -39.90, z-score = -2.03, R) >droVir3.scaffold_12963 18664260 117 - 20206255 CCAGCAACGAGCAGCAAUAAAGAGAUUAGCCUGGAGCCCUGCAAGCCAAGUGGUUUUGUUCGACAGCCGCGUCGCAACUGGCCCUGGCUGCAGGAGACCGAGCAGGAGGAGCCUGAG ((.((.....))................((.(((...((((((.((((.(.((((..(((((((......)))).))).)))))))))))))))...))).)).))........... ( -36.80, z-score = 0.71, R) >droWil1.scaffold_180772 5873931 90 + 8906247 ---------------UCCCAGAAACCUUCUAUUGAGGCCUGCAAACCAAGUGGUUUCGUGCAACAACCGCGUCGAAACUGGCCUUGGUUGCAGGAGA---CCCAUGAG--------- ---------------..........((((....))))(((((((.(((((.((((((((((.......))).)))))))...))))))))))))...---........--------- ( -30.80, z-score = -2.42, R) >droPer1.super_1 5303495 93 - 10282868 --------------ACUAAAGCCCCACU-GAUAGGGCACUGCAAGCCGAGCGGUUUCAUCCGACAGCCGCGCCGUAAUUGGCCCUGGCUGCAGGAGAGACCCCAAGAG--------- --------------......((((....-....))))...((.......))((((((.(((..((((((.((((....))))..))))))..))))))))).......--------- ( -35.90, z-score = -2.46, R) >dp4.chr4_group3 3823567 93 - 11692001 --------------ACUAAAGCCCCACU-GAUAGGGCACUGCAAGCCGAGCGGUUUCAUCCGACAGCCGCGCCGUAAUUGGCCCUGGCUGCAGGAGAGACCCCAAGAG--------- --------------......((((....-....))))...((.......))((((((.(((..((((((.((((....))))..))))))..))))))))).......--------- ( -35.90, z-score = -2.46, R) >droAna3.scaffold_12916 2512431 86 - 16180835 -------------------AGCUAUACUCCAUAGAGCCCUGCCAGCCAAGUGGUUUCACCCGCAACCCCCGCAGAAAUUGGCCAUGGCUGCAGGAGG---CCCUGGAC--------- -------------------........((((..(...(((((.(((((.((((......)))).....(((.(....))))...))))))))))...---)..)))).--------- ( -26.60, z-score = -0.02, R) >droEre2.scaffold_4845 16928275 90 + 22589142 ---------------AGCAAUUCUUUUUCCUUAGAGCCUUGCAAACCGAGUGGAUUUGUAAGACAACCACGGAGAAAUUGGCCUUGGUUGCAGGAGG---CGCAGGAA--------- ---------------...........(((((....((((((((((((....)).)))))..(.((((((.((.........)).)))))))..))))---)..)))))--------- ( -25.10, z-score = -0.39, R) >droYak2.chr2R 5128116 90 - 21139217 ---------------AGCAAAUCCUCUUUCGUAGAGCCUUGCAAACCGAGUGGAUUUGUAAGACAACCACGGAGAAAUUGGCCUUGGUUGCAGGAAG---CGCAGGAC--------- ---------------......(((((((((.......((((((((((....)).)))))))).((((((.((.........)).))))))..)))))---...)))).--------- ( -25.10, z-score = -0.58, R) >droSec1.super_7 2236982 90 - 3727775 ---------------AGCAAAUCAUCUAUGCUAGAGCCUUGCAAACCGAGUGGAUUUGUGAGACAACCACGGCGAAAUUGGCCUUGGUUGCAGGAGG---CACAGGAU--------- ---------------.........(((.((((.....((..((((((....)).))))..)).((((((.(((.......))).)))))).....))---)).)))..--------- ( -27.30, z-score = -1.12, R) >droSim1.chr2L 18302132 90 - 22036055 ---------------AGCAAAUCAUCUAUGCUAGAGCCUUGCAAACCGAGUGGAUUUGUCAGACAACCACGGCGAAAUUGGCCUUGGUUGCAGGAGG---CACAGGAU--------- ---------------.........(((.((((....(((.((((.((((((((..(((.....)))))))(((.......)))))))))))))).))---)).)))..--------- ( -26.90, z-score = -0.92, R) >consensus _______________AGCAAAUCCUCUAUCCUAGAGCCCUGCAAACCGAGUGGAUUUGUCAGACAACCACGGCGAAAUUGGCCCUGGCUGCAGGAGA___CCCAGGAG_________ ......................................((((.(((((.((((.............))))...(........).)))))))))........................ (-13.17 = -11.97 + -1.20)

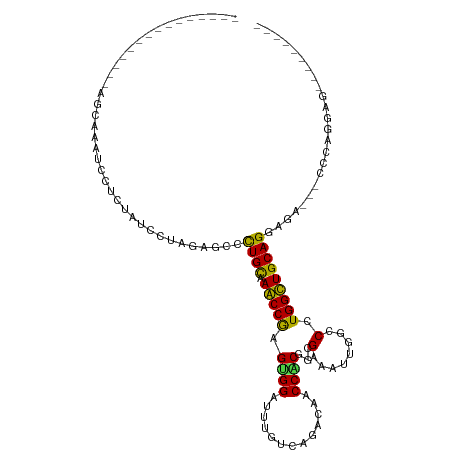
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:17 2011