| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,589,448 – 18,589,542 |
| Length | 94 |
| Max. P | 0.908121 |

| Location | 18,589,448 – 18,589,542 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.88 |
| Shannon entropy | 0.48034 |
| G+C content | 0.45621 |
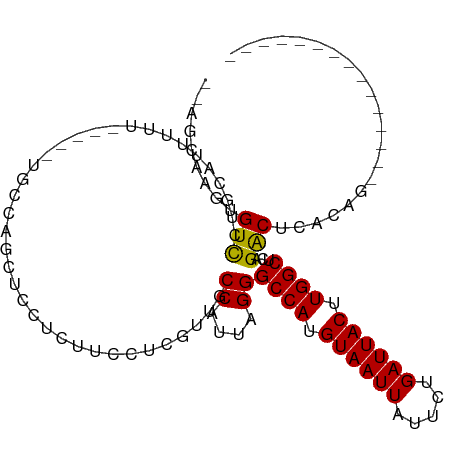
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -12.90 |
| Energy contribution | -12.56 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
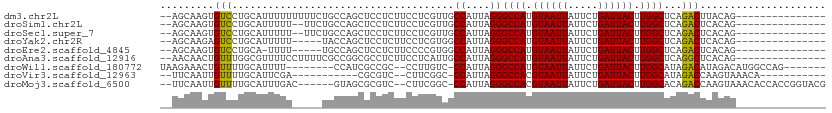
>dm3.chr2L 18589448 94 - 23011544 --AGCAAGUGUCCUGCAUUUUUUUUUCUGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUUACAG--------------- --.......(((...............((.((((...........)))).))...((((((.((((((.....)))))).)))))).)))......--------------- ( -19.00, z-score = -0.64, R) >droSim1.chr2L 18280278 92 - 22036055 --AGCAAGUGUCCUGCAUUUUU--UUCUGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACAG--------------- --.....(.(((..........--...((.((((...........)))).))...((((((.((((((.....)))))).)))))).))).)....--------------- ( -20.00, z-score = -0.83, R) >droSec1.super_7 2214682 92 - 3727775 --AGCAAGUGUCCUGCAUUUUU--UUCUGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACAG--------------- --.....(.(((..........--...((.((((...........)))).))...((((((.((((((.....)))))).)))))).))).)....--------------- ( -20.00, z-score = -0.83, R) >droYak2.chr2R 5106592 89 - 21139217 --AGCAAGAGUCCUGCAUUUUU-----UACCAGCUCCUCUUCCUCGUGGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACAG--------------- --.....(((((..........-----.....((..(........)..)).....((((((.((((((.....)))))).)))))).)))))....--------------- ( -24.10, z-score = -2.17, R) >droEre2.scaffold_4845 16906530 88 + 22589142 --AGCAAGUGUCCUGCA-UUUU-----UGCCAGCUCCUCUUCCCCGUGGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACAG--------------- --.....(((..(((..-....-----.(((((............((((((.....))))))((((((.....))))))))))).)))...)))..--------------- ( -21.80, z-score = -0.84, R) >droAna3.scaffold_12916 2492107 94 - 16180835 --AACAACUGUUUGGCGUUUUCCUUUUCGCCGGCGCCUCUUCCUCAUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGGCUCACAG--------------- --.....((((..((((((..(......)..))))))...........(((....((((((.((((((.....)))))).)))))).)))..))))--------------- ( -26.70, z-score = -2.59, R) >droWil1.scaffold_180772 5800839 93 + 8906247 UAAGAAACUGUUUUGCAUUUU--------CCAUCGCCGC--CCUUGUC-CCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCAUAGACAUAGACAUGGCCAG------- ...((((.((.....)).)))--------).......((--(..((((-((....))((((.((((((.....)))))).)))).........)))).)))...------- ( -22.90, z-score = -1.64, R) >droVir3.scaffold_12963 18636000 84 - 20206255 --UUCAAUUGUUUUGCAUUCGA-----------CGCGUC--CUUCGGC-CCAUUAGGGCCACGUAAUUAUUCUGAUUACUUGGCAUAGACCAAGUAAACA----------- --.(((......((((....((-----------....))--....(((-((....)))))..))))......)))((((((((......))))))))...----------- ( -22.90, z-score = -2.64, R) >droMoj3.scaffold_6500 26334119 100 + 32352404 --UUCAAUUGUUUUGCAUUUGAC------GUAGCGCGUC--CUUCGGC-CCAUUAGGGCCACGUAAUUAUUCUGAUUACUUGGCACAGACCAAGUAAACACCACCGGUACG --.((((.((.....)).))))(------(((.((.((.--....(((-((....)))))............((.((((((((......)))))))).))..)))).)))) ( -30.70, z-score = -3.16, R) >consensus __AGCAAGUGUCCUGCAUUUUU_____UGCCAGCUCCUCUUCCUCGUUGCCAUUAGGGCCAUGUAAUUAUUCUGAUUACUUGGCUCAGACUCACAG_______________ .........(((.....................................((....))((((.((((((.....)))))).))))...)))..................... (-12.90 = -12.56 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:15 2011