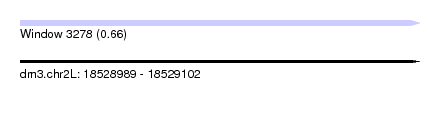
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,528,989 – 18,529,102 |
| Length | 113 |
| Max. P | 0.661266 |

| Location | 18,528,989 – 18,529,102 |
|---|---|
| Length | 113 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.89 |
| Shannon entropy | 0.74396 |
| G+C content | 0.48704 |
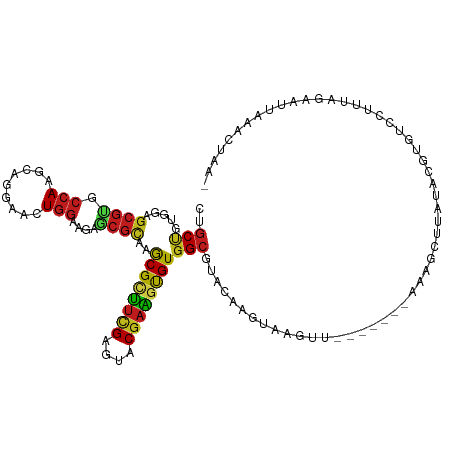
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.39 |
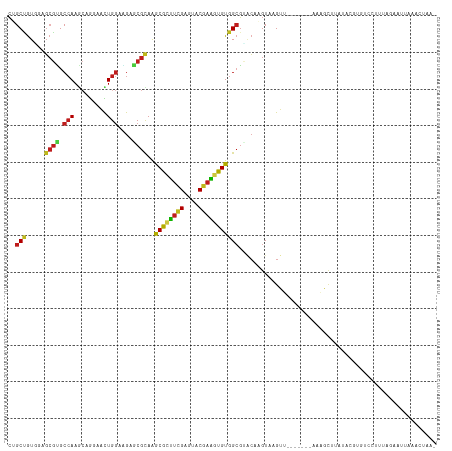
| Combinations/Pair | 1.56 |
| Mean z-score | -0.07 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
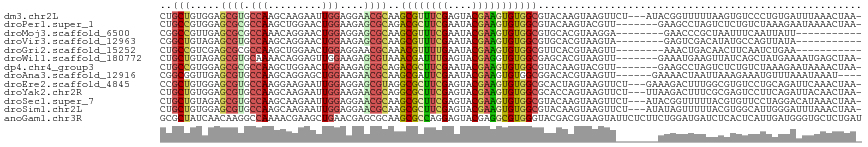
>dm3.chr2L 18528989 113 + 23011544 CUGCUGUGGAGCGUGCCAAGCAAGAAUUGGAGGAACGCAAGCGUUUCGAGUACGAAGUGUGGCGUACAAGUAAGUUCU---AUACGGUUUUUAAGUGUCCCUGUGAUUUAAACUAA- .((((.(((......))))))).((((((.(((.((((..((((((((....)))))))).))))(((..((((..((---....))..))))..))).))).)))))).......- ( -27.60, z-score = 0.31, R) >droPer1.super_1 5183948 109 + 10282868 CUGCCGUGGAGCGCGCCAAGCUGGAACUGGAAGAGCGCAGACGCUUCGAAUACGAAGUGUGGCGUACAAGUACGUU-------GAAGCCUAGUCUCUGUCUAAAGAAUAAAACUAA- ..((..(((......))).))((((...(((..((.((..((((((((....))))))))((((((....))))))-------...))))..)))...))))..............- ( -28.60, z-score = 0.45, R) >droMoj3.scaffold_6500 26242845 98 - 32352404 CGGCCGUUGAGCGCGCCAAACAGGAACUGGAGGAGCGCAAGCGUUUCGAAUACGAAGUGUGGCGUGCACGUAAGGA--------GAACCCGCUAAUUUCAAUUAUU----------- .....(((((((((((((.((.............((....))..((((....)))))).))))))))......((.--------....)).......)))))....----------- ( -28.50, z-score = -0.03, R) >droVir3.scaffold_12963 18542564 98 + 20206255 CGGCUGUAGAGCGUGCCAAGCAGGAACUGGAAGAGCGCAAGCGUUUCGAAUACGAAGUGUGGCGUGCACGUAAGUA--------GAGUCGACAUAUGCCAGUUAUA----------- .(((((.....)).))).......((((((....((....))...((((.(((...((((.....))))....)))--------...))))......))))))...----------- ( -28.20, z-score = -0.17, R) >droGri2.scaffold_15252 6493288 98 + 17193109 CUGCCGUCGAGCGCGCCAAGCUGGAACUGGAGGAACGCAAACGUUUUGAAUACGAAGUGUGGCGUUCACGUAAGUU--------AAACUGACAACUUCAAUCUGAA----------- ......((.(((.......))).))..(((((((((((..((((((((....)))))))).))))))..((.((..--------...)).))..))))).......----------- ( -23.70, z-score = 0.65, R) >droWil1.scaffold_180772 5712818 109 - 8906247 CUGCUGUAGAGCGUGCAAAACAGGAGUUGGAAGAGCGUAAACGAUUUGAGUACGAGGUGUGGCGAGCACGUAAGUU-------GAAAUGAAGUUAUCAGCUAUGAAAAUGAGCUAA- ..(((.....((((((....((.(..(((...(((((....)).))).....)))..).))....)))))).((((-------((.(......).)))))).........)))...- ( -20.70, z-score = 1.00, R) >dp4.chr4_group3 3702702 109 + 11692001 CUGCCGUGGAGCGCGCCAAGCUGGAACUGGAAGAGCGCAGACGCUUCGAAUACGAAGUGUGGCGUACAAGUACGUU-------GAAGCCUAGUCUCUGUCUAAAGAAUAAAACUAA- ..((..(((......))).))((((...(((..((.((..((((((((....))))))))((((((....))))))-------...))))..)))...))))..............- ( -28.60, z-score = 0.45, R) >droAna3.scaffold_12916 2437282 107 + 16180835 CGGCGGUUGAGCGUGCCAAGCAGGAGCUGGAAGAACGCAAGCGAUUCGAAUACGAAGUGUGGCGGACACGUAAGUU------GAAAACUAAUUAAAGAAAUGUUUAAAUAAAU---- ((((......((((((((.((....))))).....(((..(((.((((....)))).))).)))..)))))..)))------)..............................---- ( -19.90, z-score = 0.75, R) >droEre2.scaffold_4845 16847276 113 - 22589142 CCGCUGUGGAGCGUGCCAAGGAAGAAUUGGAGGAGCGUAGGCGCUUCGAGUACGAAGUGUGGCGCACUAGUAAGUUCU---GAAAGACUUUGGCGUGUCCUGCAGAUUCAAACUAA- ...(((..(..((((((((((.((((((..((..((((..((((((((....)))))))).)))).))....))))))---......))))))))))..)..)))...........- ( -43.70, z-score = -2.86, R) >droYak2.chr2R 5046505 113 + 21139217 CUGCUGUGGAGCGUGCCAAGCAAGAAUUGGAAGAACGCAGGCGCUUCGAGUACGAAGUGUGGCGCACCAGUAAGUUCU---UUAAGACUUUCGCGAGUCCUUCAGAUUACAACUAA- ...(((.(((.((((((((.......))))((((((((..((((((((....)))))))).))((....))..)))))---).........))))..)))..)))...........- ( -31.60, z-score = 0.24, R) >droSec1.super_7 2155038 113 + 3727775 CUGCUGUAGAGCGUGCCAAGCAAGAAUUGGAGGAACGCAAGCGCUUCGAGUACGAAGUGUGGCGUACAAGUAAGUUCU---AUACGGUUUUUACGUGUUCCUAGGACAUAAACUAA- (((.((((((((...((((.......))))..(.((((..((((((((....)))))))).)))).)......)))))---)))))).......(((((.....))))).......- ( -33.30, z-score = -1.10, R) >droSim1.chr2L 18219746 113 + 22036055 CUGCUGUGGAGCGUGCCAAGCAAGAAUUGGAGGAACGCAAGCGCUUCGAGUACGAAGUGUGGCGUACAAGUAAGUUCU---AUAUAGUUUUUACGUGGCAUUGGGAUUUAAACUAA- ..(((....)))((((((.(.((((((((..(((((((..((((((((....)))))))).)).((....)).)))))---...)))))))).).))))))(((........))).- ( -32.00, z-score = -0.90, R) >anoGam1.chr3R 9728631 117 + 53272125 GCGCUAUCAACAAGGCCAAAACGAAGCUGAACGAGCGCAAGCGCCAGGAGUACGAGGCGUGGGUACGACGUAAGUAUUCUCUUCUGGAUGAUCUCACUCAUUGAUGGGUGCUCUGAU (((((........(((.........))).....))))).((((((.(((((((...((((.(...).))))..)))))))......(((((......)))))....))))))..... ( -33.22, z-score = 0.31, R) >consensus CUGCUGUGGAGCGUGCCAAGCAGGAACUGGAAGAGCGCAAGCGCUUCGAGUACGAAGUGUGGCGUACAAGUAAGUU_______AAAGCUUAUACGUGUCCUUUAGAAUUAAACUAA_ ..(((.....((((.(((.........)))....))))..((((((((....)))))))))))...................................................... (-16.62 = -16.22 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:10 2011