| Sequence ID | dm3.chr2L |
|---|---|
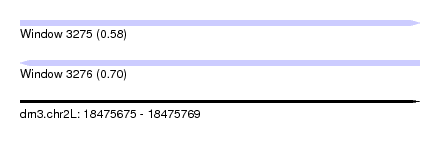
| Location | 18,475,675 – 18,475,769 |
| Length | 94 |
| Max. P | 0.700227 |

| Location | 18,475,675 – 18,475,769 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 63.19 |
| Shannon entropy | 0.72878 |
| G+C content | 0.57603 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -8.44 |
| Energy contribution | -9.74 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.82 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
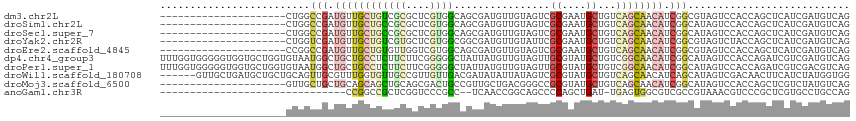
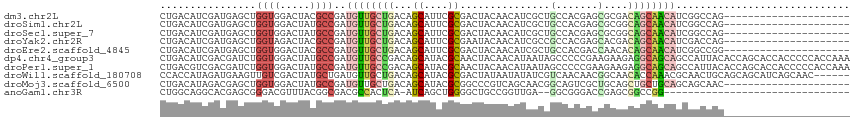
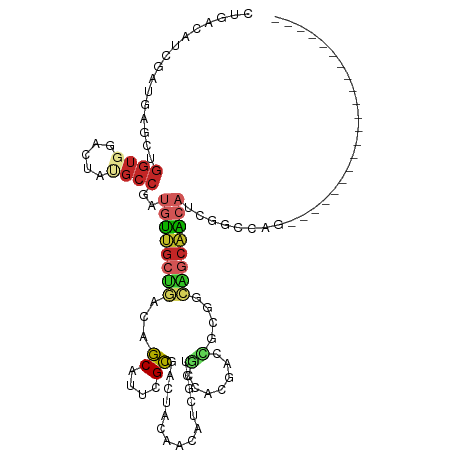
>dm3.chr2L 18475675 94 + 23011544 ---------------------CUGGCCGAUGUUGCUGUCGCGCUCGUGGCAGCGAUGUUGUAGUCGCGAAUGCUGUCAGCAACAUCGGCGUAGUCCACCAGCUCAUCGAUGUCAG ---------------------((((((((((((((((.((((.(((((((.(((....))).))))))).))).).)))))))))))))(.(((......))))........))) ( -42.80, z-score = -3.31, R) >droSim1.chr2L 18164747 94 + 22036055 ---------------------CUGGCCGAUGUUGCUGCCGCGCUCGUGGCAGCGAUGUUGUAGUCGCGAAUGCUGUCAGCAACAUCGGCAUAGUCCACCAGCUCAUCGAUGUCAG ---------------------(((((((((((((((((.(((.(((((((.(((....))).))))))).))).).))))))))))))).)))...................... ( -42.00, z-score = -3.00, R) >droSec1.super_7 2101542 94 + 3727775 ---------------------CUGGCCGAUGUUGCUGCCGCGCUCGUGGCAGCGAUGUUGUAGUCGCGAAUGCUGUCAGCAACAUCGGCAUAGUCCACCAGCUCAUCGAUGUCAG ---------------------(((((((((((((((((.(((.(((((((.(((....))).))))))).))).).))))))))))))).)))...................... ( -42.00, z-score = -3.00, R) >droYak2.chr2R 4992132 94 + 21139217 ---------------------CUGGUCGAUGUUGCUGUCGUGCUCGUGGCGGCGAUGUUGUAUUCGCGAAUGCUGUCAGCAACAUCGGCGUAGUCUACCAGCUCAUCGAUGUCAG ---------------------((((((((((..((((..(((..(...(((.(((((((((...(((....)).)...))))))))).))).)..))))))).)))))))..))) ( -35.70, z-score = -1.64, R) >droEre2.scaffold_4845 16793493 94 - 22589142 ---------------------CCGGCCGAUGUUGCUGUGUUGGUCGUGGCAGCGAUGUUGUAGUCGCGAAUGCUGUCAGCAACAUCGGCGUAGUCCACCAGCUCAUCGAUGUCAG ---------------------.(((((((((((((((.((...(((((((.(((....))).)))))))..))...)))))))))))))(.(((......))))..))....... ( -39.30, z-score = -2.69, R) >dp4.chr4_group3 3638872 115 + 11692001 UUUGGUGGGGGUGGUGCUGGUGUAAUGGCUGCUGCCUCUUCUUCGGGGGCUAUUAUGUUGUAGUUGCGUAUGCUGUCGGCAACAUCGGCAUAGUCCACCAGAUCGUCGAUGUCAG (((((((((....((((((((((..((((.((.((((((.....))))))...(((((.......))))).)).))))...))))))))))..)))))))))............. ( -43.10, z-score = -1.88, R) >droPer1.super_1 5120137 115 + 10282868 UUUGGUGGGGGUGGUGCUGGUGUAAUGGCUGCUGCCUCUUCUUCGGGGGCUAUUAUGUUGUAGUUGCGUAUGCUGUCGGCAACAUCGGCAUAGUCCACCAGAUCGUCGACGUCAG (((((((((....((((((((((..((((.((.((((((.....))))))...(((((.......))))).)).))))...))))))))))..)))))))))............. ( -43.10, z-score = -1.71, R) >droWil1.scaffold_180708 11628310 109 + 12563649 ------GUUGCUGAUGCUGCUGCAGUUGCGUUUGGUGUUGCCGUUGUUGACGAUAUAUUAUAGUCGCGUAUGCUGUCAGCAACAUCAGCAUAGUCGACAACUUCAUCUAUGGUGG ------...(((((((.((((((((((((((..((.....))......(((...........)))))))).)))).))))).)))))))((((..((.....))..))))..... ( -35.80, z-score = -0.96, R) >droMoj3.scaffold_6500 26171777 94 - 32352404 ---------------------GUUGCUGCUGCAGCAGCUGCAGCGACUGCCGUUGCUGACGGGCCGCGUAUGCUGUCAGCAACAUCGGCAUAGUCCACCAGCUCGUCUAUGUCAG ---------------------((((((((.((....)).))))))))....(((((((((((..........)))))))))))...(((((((.(.........).))))))).. ( -41.60, z-score = -1.81, R) >anoGam1.chr3R 10314758 81 + 53272125 -------------------------------CCGGCCGCUCGGUCCCGCC--UCAACCGGCAGCCCCAGCUGAU-UGAGUGGCGUCGCCGUAAACGUCCCGCUCGUGCCUGCCAG -------------------------------..(((.((.((((..((((--((((.((((.......)))).)-)))..))))..))))...(((.......)))))..))).. ( -27.80, z-score = -0.37, R) >consensus _____________________CUGGCCGAUGUUGCUGCCGCGCUCGUGGCAGCGAUGUUGUAGUCGCGAAUGCUGUCAGCAACAUCGGCAUAGUCCACCAGCUCAUCGAUGUCAG .........................(((.((((((((((((....))))................((....))...)))))))).)))........................... ( -8.44 = -9.74 + 1.30)
| Location | 18,475,675 – 18,475,769 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 63.19 |
| Shannon entropy | 0.72878 |
| G+C content | 0.57603 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -7.63 |
| Energy contribution | -8.10 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
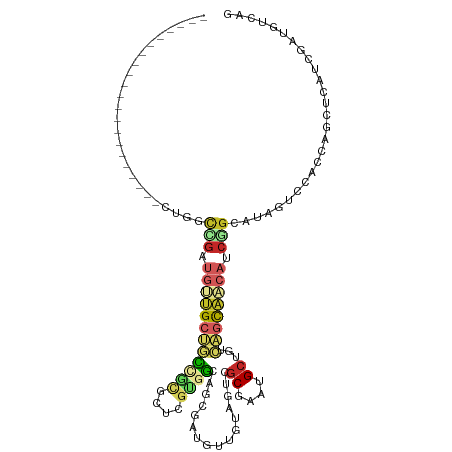
>dm3.chr2L 18475675 94 - 23011544 CUGACAUCGAUGAGCUGGUGGACUACGCCGAUGUUGCUGACAGCAUUCGCGACUACAACAUCGCUGCCACGAGCGCGACAGCAACAUCGGCCAG--------------------- (((.(((((......)))))......(((((((((((((...((....((((........)))).((.....))))..))))))))))))))))--------------------- ( -37.20, z-score = -3.30, R) >droSim1.chr2L 18164747 94 - 22036055 CUGACAUCGAUGAGCUGGUGGACUAUGCCGAUGUUGCUGACAGCAUUCGCGACUACAACAUCGCUGCCACGAGCGCGGCAGCAACAUCGGCCAG--------------------- (((.(((((......)))))......(((((((((((((...((....((((........)))).((.....))))..))))))))))))))))--------------------- ( -38.00, z-score = -2.76, R) >droSec1.super_7 2101542 94 - 3727775 CUGACAUCGAUGAGCUGGUGGACUAUGCCGAUGUUGCUGACAGCAUUCGCGACUACAACAUCGCUGCCACGAGCGCGGCAGCAACAUCGGCCAG--------------------- (((.(((((......)))))......(((((((((((((...((....((((........)))).((.....))))..))))))))))))))))--------------------- ( -38.00, z-score = -2.76, R) >droYak2.chr2R 4992132 94 - 21139217 CUGACAUCGAUGAGCUGGUAGACUACGCCGAUGUUGCUGACAGCAUUCGCGAAUACAACAUCGCCGCCACGAGCACGACAGCAACAUCGACCAG--------------------- (((...((((((.(((((((.....(((.(((((((....))))))).)))..)))....(((..((.....)).)))))))..)))))).)))--------------------- ( -27.50, z-score = -1.92, R) >droEre2.scaffold_4845 16793493 94 + 22589142 CUGACAUCGAUGAGCUGGUGGACUACGCCGAUGUUGCUGACAGCAUUCGCGACUACAACAUCGCUGCCACGACCAACACAGCAACAUCGGCCGG--------------------- (((.(((((......)))))......(((((((((((((...(((...((((........)))))))...(.....).))))))))))))))))--------------------- ( -32.20, z-score = -2.75, R) >dp4.chr4_group3 3638872 115 - 11692001 CUGACAUCGACGAUCUGGUGGACUAUGCCGAUGUUGCCGACAGCAUACGCAACUACAACAUAAUAGCCCCCGAAGAAGAGGCAGCAGCCAUUACACCAGCACCACCCCCACCAAA ...............((((((....(((..((((((....))))))..)))............................(((....)))..................)))))).. ( -22.50, z-score = -0.71, R) >droPer1.super_1 5120137 115 - 10282868 CUGACGUCGACGAUCUGGUGGACUAUGCCGAUGUUGCCGACAGCAUACGCAACUACAACAUAAUAGCCCCCGAAGAAGAGGCAGCAGCCAUUACACCAGCACCACCCCCACCAAA ..((((....)).))((((((....(((..((((((....))))))..)))............................(((....)))..................)))))).. ( -23.20, z-score = -0.50, R) >droWil1.scaffold_180708 11628310 109 - 12563649 CCACCAUAGAUGAAGUUGUCGACUAUGCUGAUGUUGCUGACAGCAUACGCGACUAUAAUAUAUCGUCAACAACGGCAACACCAAACGCAACUGCAGCAGCAUCAGCAAC------ .....((((.(((.....))).))))(((((((((((((.(((.....(((((.(((...))).)))......(....).......))..))))))))))))))))...------ ( -34.00, z-score = -3.58, R) >droMoj3.scaffold_6500 26171777 94 + 32352404 CUGACAUAGACGAGCUGGUGGACUAUGCCGAUGUUGCUGACAGCAUACGCGGCCCGUCAGCAACGGCAGUCGCUGCAGCUGCUGCAGCAGCAAC--------------------- .............(((....((((..(((...(((((((((.((.......))..))))))))))))))))(((((((...))))))))))...--------------------- ( -38.00, z-score = -0.92, R) >anoGam1.chr3R 10314758 81 - 53272125 CUGGCAGGCACGAGCGGGACGUUUACGGCGACGCCACUCA-AUCAGCUGGGGCUGCCGGUUGA--GGCGGGACCGAGCGGCCGG------------------------------- (((((......(((.(((.(((.....))).).)).))).-....(((.((.(((((......--)))))..)).))).)))))------------------------------- ( -34.00, z-score = -0.52, R) >consensus CUGACAUCGAUGAGCUGGUGGACUAUGCCGAUGUUGCUGACAGCAUUCGCGACUACAACAUCGCUGCCACGACCGCGGCAGCAACAUCGGCCAG_____________________ ................((((.....))))..((((((((...((....))...............(.......)....))))))))............................. ( -7.63 = -8.10 + 0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:08 2011